2IG3
 
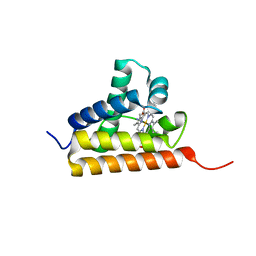 | | Crystal structure of group III truncated hemoglobin from Campylobacter jejuni | | Descriptor: | ACETATE ION, CYANIDE ION, Group III truncated haemoglobin, ... | | Authors: | Nardini, M, Pesce, A, Labarre, M, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural determinants in the group III truncated hemoglobin from Campylobacter jejuni.
J.Biol.Chem., 281, 2006
|
|
3ZX1
 
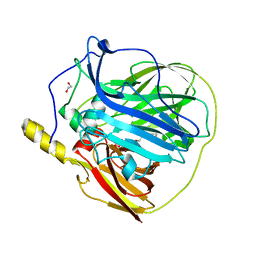 | | Multicopper oxidase from Campylobacter jejuni: a metallo-oxidase | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, OXIDOREDUCTASE, ... | | Authors: | Silva, C.S, Durao, P, Fillat, A, Lindley, P.F, Martins, L.O, Bento, I. | | Deposit date: | 2011-08-04 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Multicopper Oxidase from the Pathogenic Bacterium Campylobacter Jejuni Cgug11284: Characterization of a Metallo-Oxidase.
Metallomics, 4, 2012
|
|
6XJ7
 
 | |
6XJ6
 
 | |
2QCO
 
 | | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni | | Descriptor: | CmeR, GLYCEROL | | Authors: | Gu, R, Su, C, Shi, F, Li, M, McDermott, G, Zhang, Q, Yu, E.W. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni.
J.Mol.Biol., 372, 2007
|
|
2P56
 
 | | Crystal structure of alpha-2,3-sialyltransferase from Campylobacter jejuni in apo form | | Descriptor: | 1,2-ETHANEDIOL, Alpha-2,3-sialyltransferase | | Authors: | Chiu, C.P, Lairson, L.L, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2007-03-14 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of the alpha-2,3-Sialyltransferase Cst-I from Campylobacter jejuni in Apo and Substrate-Analogue Bound Forms.
Biochemistry, 46, 2007
|
|
2HXW
 
 | | Crystal Structure of Peb3 from Campylobacter jejuni | | Descriptor: | CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Rangarajan, E.S, Bhatia, S, Watson, D.C, Munger, C, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-08-04 | | Release date: | 2007-05-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural context for protein N-glycosylation in bacteria: The structure of PEB3, an adhesin from Campylobacter jejuni.
Protein Sci., 16, 2007
|
|
2P2V
 
 | | Crystal structure analysis of monofunctional alpha-2,3-sialyltransferase Cst-I from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, Alpha-2,3-sialyltransferase, CHLORIDE ION, ... | | Authors: | Chiu, C.P, Lairson, L.L, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2007-03-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of the alpha-2,3-Sialyltransferase Cst-I from Campylobacter jejuni in Apo and Substrate-Analogue Bound Forms.
Biochemistry, 46, 2007
|
|
3D6X
 
 | | Crystal structure of Campylobacter jejuni FabZ | | Descriptor: | (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase | | Authors: | Yokoyama, T, Yeo, H.J. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Campylobacter jejuni fatty acid synthase II: structural and functional analysis of beta-hydroxyacyl-ACP dehydratase (FabZ).
Biochem.Biophys.Res.Commun., 380, 2009
|
|
6X80
 
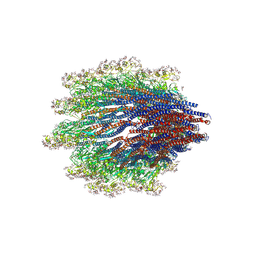 | | Structure of the Campylobacter jejuni G508A Flagellar Filament | | Descriptor: | 5,7-diamino-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid, Flagellin A | | Authors: | Kreutzberger, M.A.B, Wang, F, Egelman, E.H. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structure of the Campylobacter jejuni flagellar filament reveals how epsilon Proteobacteria escaped Toll-like receptor 5 surveillance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Z05
 
 | |
3D6L
 
 | |
3HBM
 
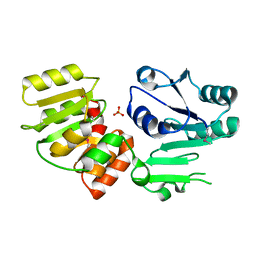 | | Crystal Structure of PseG from Campylobacter jejuni | | Descriptor: | SULFATE ION, UDP-sugar hydrolase | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
3FIR
 
 | | Crystal structure of Glycosylated K135E PEB3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-L-glucopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-12 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
4ETS
 
 | |
3TPF
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ornithine carbamoyltransferase | | Authors: | Shabalin, I.G, Onopriyenko, O, Grimshaw, S, Porebski, P.J, Grabowski, M, Savchenko, A, Chruszcz, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni at 2.7 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6KF3
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF4
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
2R9S
 
 | | c-Jun N-terminal Kinase 3 with 3,5-Disubstituted Quinoline inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 10, N-(tert-butyl)-4-[5-(pyridin-2-ylamino)quinolin-3-yl]benzenesulfonamide, ... | | Authors: | Habel, J. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3,5-Disubstituted quinolines as novel c-Jun N-terminal kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2D0D
 
 | | Crystal Structure of a Meta-cleavage Product Hydrolase (CumD) A129V Mutant | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, CHLORIDE ION, PHOSPHATE ION | | Authors: | Jun, S.Y, Fushinobu, S, Nojiri, H, Omori, T, Shoun, H, Wakagi, T. | | Deposit date: | 2005-08-01 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Improving the catalytic efficiency of a meta-cleavage product hydrolase (CumD) from Pseudomonas fluorescens IP01
Biochim.Biophys.Acta, 1764, 2006
|
|
5DQP
 
 | | EDTA monooxygenase (EmoA) from Chelativorans sp. BNC1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, EDTA monooxygenase, SULFATE ION | | Authors: | Jun, S.Y, Youn, B, Xun, L, Kang, C, Lewis, K.M. | | Deposit date: | 2015-09-15 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Structural and biochemical characterization of EDTA monooxygenase and its physical interaction with a partner flavin reductase.
Mol.Microbiol., 100, 2016
|
|
6KF9
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*CP*GP*GP*TP*AP*AP*TP*CP*AP*CP*GP*CP*TP*CP*C)-3'), DNA-directed RNA polymerase subunit, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
4QIW
 
 | |
6AT7
 
 | | Phenylalanine Ammonia-Lyase (PAL) from Sorghum bicolor | | Descriptor: | AMMONIUM ION, Phenylalanine ammonia-lyase | | Authors: | Jun, S.Y, Kang, C. | | Deposit date: | 2017-08-28 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Biochemical and Structural Analysis of Substrate Specificity of a Phenylalanine Ammonia-Lyase.
Plant Physiol., 176, 2018
|
|
4IZY
 
 | |
