3EUX
 
 | | Crystal Structure of Crosslinked Ribonuclease A | | Descriptor: | Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV4
 
 | | Crystal Structure of Ribonuclease A in 50% Trifluoroethanol | | Descriptor: | Ribonuclease pancreatic, TRIFLUOROETHANOL | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EUZ
 
 | | Crystal Structure of Ribonuclease A in 50% Dimethylformamide | | Descriptor: | DIMETHYLFORMAMIDE, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV1
 
 | | Crystal Structure of Ribonuclease A in 70% Hexanediol | | Descriptor: | HEXANE-1,6-DIOL, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV2
 
 | | Crystal Structure of Ribonuclease A in 70% Isopropanol | | Descriptor: | ISOPROPYL ALCOHOL, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EV5
 
 | | Crystal Structure of Ribonuclease A in 1M Trimethylamine N-Oxide | | Descriptor: | Ribonuclease pancreatic, trimethylamine oxide | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
7PH1
 
 | | Trypsin in complex with BPTI mutant (2S)-2-amino-4-monofluorobutanoic acid | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Loll, B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Water Network in the Binding Pocket of Fluorinated BPTI-Trypsin Complexes─Insights from Simulation and Experiment.
J.Phys.Chem.B, 126, 2022
|
|
9GLY
 
 | |
4WEP
 
 | | Apo YehZ from Escerichia coli | | Descriptor: | Putative osmoprotectant uptake system substrate-binding protein OsmF | | Authors: | Kimber, M.S, Lang, S, Mendoza, K, Wood, J.M. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | YehZYXW of Escherichia coli Is a Low-Affinity, Non-Osmoregulatory Betaine-Specific ABC Transporter.
Biochemistry, 54, 2015
|
|
7O07
 
 | |
4WJX
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain at 1.0 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WEV
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and sulindac | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Crespo, I, Porte, S, Pares, X, Farres, J, Podjarny, A. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural analysis of sulindac as an inhibitor of aldose reductase and AKR1B10.
Chem.Biol.Interact., 234, 2015
|
|
4WKA
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6TO0
 
 | |
4WU0
 
 | | Structural Analysis of C. acetobutylicum ATCC 824 Glycoside Hydrolase From Family 105 | | Descriptor: | Similar to yteR (Bacilus subtilis) | | Authors: | Germane, K.L, Servinsky, M.D, Gerlach, E.S, Sund, C.J, Hurley, M.M. | | Deposit date: | 2014-10-30 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of Clostridium acetobutylicum ATCC 824 glycoside hydrolase from CAZy family GH105.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6TOW
 
 | |
7PJD
 
 | |
6TRH
 
 | |
4WAK
 
 | |
4WAM
 
 | |
9FWW
 
 | | Human NKp30 in complex with a VHH variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Natural cytotoxicity triggering receptor 3, VHH, ... | | Authors: | Musil, D, Freire, F. | | Deposit date: | 2024-07-01 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | On the humanization of VHHs: Prospective case studies, experimental and computational characterization of structural determinants for functionality.
Protein Sci., 33, 2024
|
|
3DV2
 
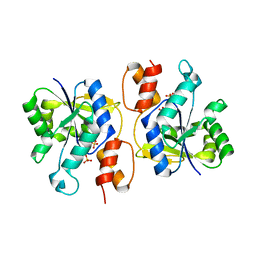 | | Crystal Structure of nicotinic acid mononucleotide adenylyltransferase from Bacillus anthracis | | Descriptor: | Nicotinate (Nicotinamide) nucleotide adenylyltransferase, SULFATE ION | | Authors: | Lu, S, Smith, C.D, Yang, Z, Pruett, P.S, Nagy, L, McCombs, D.P, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of nicotinic acid mononucleotide adenylyltransferase from Bacillus anthracis.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
7P8O
 
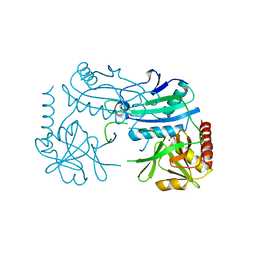 | | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its intermediate form | | Descriptor: | Aminotransferase class IV, MAGNESIUM ION, SULFATE ION | | Authors: | Matyuta, I.O, Boyko, K.M, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Incorporation of pyridoxal-5'-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
1FYU
 
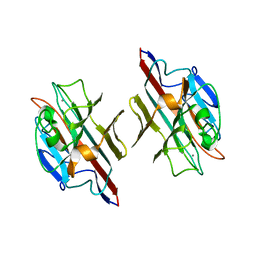 | |
3FSJ
 
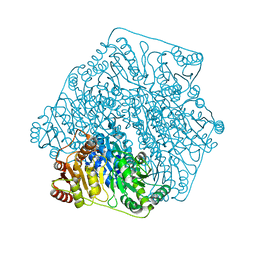 | | Crystal structure of benzoylformate decarboxylase in complex with the inhibitor MBP | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, CALCIUM ION | | Authors: | Brandt, G.S, Kenyon, G.L, McLeish, M.J, Jordan, F, Petsko, G.A, Ringe, D. | | Deposit date: | 2009-01-09 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Snapshot of a reaction intermediate: analysis of benzoylformate decarboxylase in complex with a benzoylphosphonate inhibitor.
Biochemistry, 48, 2009
|
|
