1GMM
 
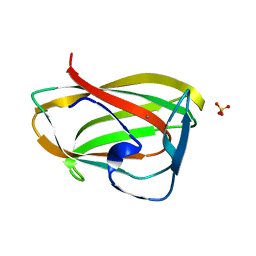 | | Carbohydrate binding module CBM6 from xylanase U Clostridium thermocellum | | Descriptor: | CALCIUM ION, CBM6, SODIUM ION, ... | | Authors: | Czjzek, M, Mosbah, A, Bolam, D, Allouch, J, Zamboni, V, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2001-09-19 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Location of the Ligand-Binding Site of Carbohydrate-Binding Modules that Have Evolved from a Common Sequence is not Conserved.
J.Biol.Chem., 276, 2001
|
|
2V4V
 
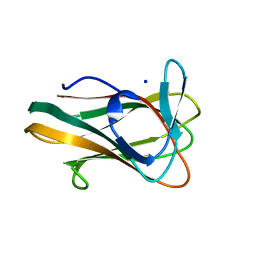 | | Crystal Structure of a Family 6 Carbohydrate-Binding Module from Clostridium cellulolyticum in complex with xylose | | Descriptor: | GH59 GALACTOSIDASE, SODIUM ION, beta-D-xylopyranose | | Authors: | Abbott, D.W, Ficko-Blean, E, Lammerts van Bueren, A, Coutinho, P.M, Henrissat, B, Gilbert, H.J, Boraston, A.B. | | Deposit date: | 2008-09-29 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the Structural and Functional Diversity of Plant Cell Wall Specific Family 6 Carbohydrate Binding Modules.
Biochemistry, 48, 2009
|
|
2W46
 
 | | CBM35 from Cellvibrio japonicus Abf62 | | Descriptor: | CALCIUM ION, ESTERASE D, SODIUM ION | | Authors: | Montainer, C, Lammerts van Bueren, A, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W87
 
 | | Xyl-CBM35 in complex with glucuronic acid containing disaccharide. | | Descriptor: | CALCIUM ION, ESTERASE D, UNKNOWN LIGAND, ... | | Authors: | Montainer, C, Bueren, A.L.v, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1O8P
 
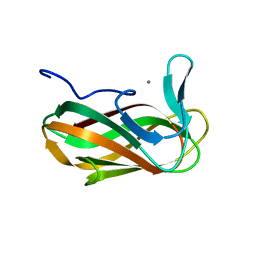 | | Unbound structure of CsCBM6-3 from Clostridium stercorarium | | Descriptor: | CALCIUM ION, PUTATUVE ENDO-XYLANASE | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilbrun, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains.
J. Mol. Biol., 327, 2003
|
|
1O8S
 
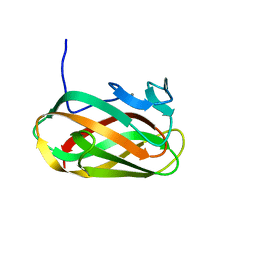 | | Structure of CsCBM6-3 from Clostridium stercorarium in complex with cellobiose | | Descriptor: | CALCIUM ION, PUTATIVE ENDO-XYLANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilbrun, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Ligand Binding of Carbohydrate-Binding Module Cscbm6-3 Reveals Similarities with Fucose-Specific Lectins and Galactose-Binding Domains
J.Mol.Biol., 327, 2003
|
|
2CDO
 
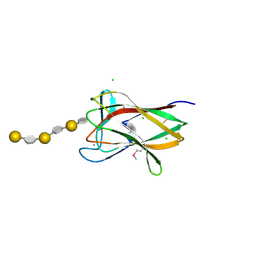 | | structure of agarase carbohydrate binding module in complex with neoagarohexaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE 1, ... | | Authors: | Henshaw, J, Horne, A, Van Bueren, A.L, Money, V.A, Bolam, D.N, Czjzek, M, Weiner, R.M, Hutcheson, S.W, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2006-01-25 | | Release date: | 2006-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Family 6 Carbohydrate Binding Modules in Beta-Agarases Display Exquisite Selectivity for the Non- Reducing Termini of Agarose Chains.
J.Biol.Chem., 281, 2006
|
|
1W0N
 
 | | Structure of uncomplexed Carbohydrate Binding Domain CBM36 | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, MAGNESIUM ION, ... | | Authors: | Jamal, S, Boraston, A.B, Davies, G.J. | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
1W9T
 
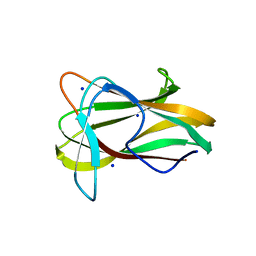 | | Structure of a beta-1,3-glucan binding CBM6 from Bacillus halodurans in complex with xylobiose | | Descriptor: | BH0236 PROTEIN, SODIUM ION, alpha-D-xylopyranose, ... | | Authors: | Boraston, A.B, van Bueren, A.L. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Family 6 Carbohydrate Binding Modules Recognize the Non-Reducing End of Beta-1,3-Linked Glucans by Presenting a Unique Ligand Binding Surface
J.Biol.Chem., 280, 2005
|
|
1W9S
 
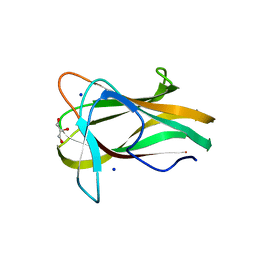 | |
2CDP
 
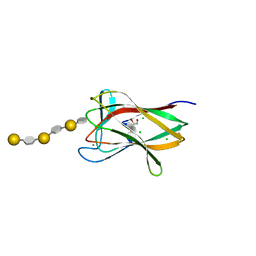 | | Structure of a CBM6 in complex with neoagarohexaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE 1, ... | | Authors: | Henshaw, J, Horne, A, Van Bueren, A.L, Money, V.A, Bolam, D.N, Czjzek, M, Weiner, R.M, Hutcheson, S.W, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2006-01-26 | | Release date: | 2006-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Family 6 Carbohydrate Binding Modules in Beta-Agarases Display Exquisite Selectivity for the Non- Reducing Termini of Agarose Chains.
J.Biol.Chem., 281, 2006
|
|
1W9W
 
 | |
4QB6
 
 | | Structure of CBM35 in complex with aldouronic acid | | Descriptor: | CALCIUM ION, Xyn30D, alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2014-05-06 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Analysis of Glucuronoxylan-specific Xyn30D and Its Attached CBM35 Domain Gives Insights into the Role of Modularity in Specificity.
J.Biol.Chem., 289, 2014
|
|
4QB1
 
 | |
4QB2
 
 | | Structure of CBM35 in complex with glucuronic acid | | Descriptor: | CALCIUM ION, Xyn30D, alpha-D-glucopyranuronic acid, ... | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2014-05-06 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Glucuronoxylan-specific Xyn30D and Its Attached CBM35 Domain Gives Insights into the Role of Modularity in Specificity.
J.Biol.Chem., 289, 2014
|
|
4QAW
 
 | |
1OD3
 
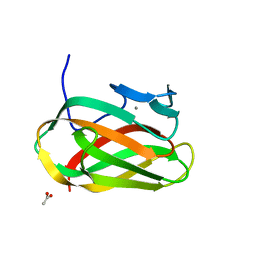 | | Structure of CSCBM6-3 From Clostridium stercorarium in complex with laminaribiose | | Descriptor: | ACETIC ACID, CALCIUM ION, PUTATIVE XYLANASE, ... | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure and Ligand Binding of Carbohydrate-Binding Module Cscbm6-3 Reveals Similarities with Fucose-Specific Lectins and Galactose-Binding Domains
J.Mol.Biol., 327, 2003
|
|
1NAE
 
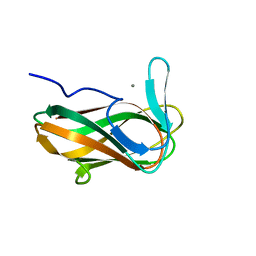 | | Structure of CsCBM6-3 from Clostridium stercorarium in complex with xylotriose | | Descriptor: | CALCIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, putative xylanase | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G. | | Deposit date: | 2002-11-27 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains
J.Mol.Biol., 327, 2003
|
|
5FUI
 
 | | Crystal structure of the C-terminal CBM6 of LamC a marine laminarianse from Zobellia galactanivorans | | Descriptor: | 2-AMINOMETHYL-PYRIDINE, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jam, M, Legentil, L, Sylla, B, Hehemann, J.H, Ficko-Blean, E, Ferrieres, V, Czjzek, M, Michel, G. | | Deposit date: | 2016-01-27 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unraveling the Multivalent Binding of a Marine Family 6 Carbohydrate-Binding Module with its Native Laminarin Ligand.
FEBS J., 283, 2016
|
|
1UX7
 
 | | Carbohydrate-Binding Module CBM36 in complex with calcium and xylotriose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, SULFATE ION, ... | | Authors: | Davies, G.J, Boraston, A.B, Jamal, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
1UY0
 
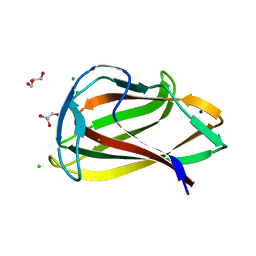 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with glc-1,3-glc-1,4-glc-1,3-glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYX
 
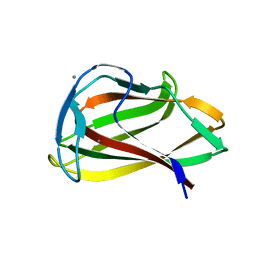 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellobiose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYZ
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with xylotetraose | | Descriptor: | CALCIUM ION, CELLULASE B, GLYCEROL, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UY4
 
 | |
1UZ0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with Glc-4Glc-3Glc-4Glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
