2CXC
 
 | | Crystal structure of archaeal transcription termination factor NusA | | Descriptor: | NusA | | Authors: | Shibata, R, Bessho, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and RNA-binding analysis of the archaeal transcription factor NusA
Biochem.Biophys.Res.Commun., 355, 2007
|
|
2CY1
 
 | | Crystal structure of APE1850 | | Descriptor: | NusA protein homolog | | Authors: | Shibata, R, Bessho, Y, Umehara, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of the archaeal transcription termination factor NusA: a significant decrease in twinning under microgravity conditions
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1WH9
 
 | | Solution structure of the KH domain of human ribosomal protein S3 | | Descriptor: | 40S ribosomal protein S3 | | Authors: | Nameki, N, Tomizawa, T, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KH domain of human ribosomal protein S3
To be Published
|
|
5OT7
 
 | | Elongation factor G-ribosome complex captures in the absence of inhibitors. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mace, K, Giudice, E, Chat, S, Gillet, R. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-14 | | Last modified: | 2018-04-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structure of an elongation factor G-ribosome complex captured in the absence of inhibitors.
Nucleic Acids Res., 46, 2018
|
|
5BR8
 
 | | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sierra, R.G, Gati, C, Laksmono, H, Dao, E.H, Gul, S, Fuller, F, Kern, J, Chatterjee, R, Ibrahim, M, Brewster, A, Young, I.D, Michels-Clark, T, Aquila, A, Mengning, L, Hunter, M.S, Koglin, J.E, Boutet, S, Junco, E.A, Hayes, B, Bogan, M.J, Hampton, C.Y, Puglisi, E.V, Sauter, N.K, Stan, C.A, Zouni, A, Yano, J, Yachandra, V.K, Soltis, S.M, Puglisi, J.D, DeMirci, H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin
To Be Published
|
|
3T1Y
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA Lysine 3 (TRNALYS3) bound to an mRNA with an AAG-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
2HHH
 
 | | Crystal structure of kasugamycin bound to the 30S ribosomal subunit | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schluenzen, F. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2F4V
 
 | | 30S ribosome + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-{(1R,2S,3R,4R,5S)-5-AMINO-2-{2-[(2-AMINOETHYL)AMINO]ETHOXY}-4-[(2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-3-HYDROXYCYCLOHEXYL}-2-HYDROXYBUTANAMIDE, (2S,4S,4AR,5AS,6S,11R,11AS,12R,12AR)-7-CHLORO-4-(DIMETHYLAMINO)-6,10,11,12-TETRAHYDROXY-1,3-DIOXO-1,2,3,4,4A,5,5A,6,11,11A,12,12A-DODECAHYDROTETRACENE-2-CARBOXAMIDE, 16S ribosomal RNA, ... | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosome aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
3T1H
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA lysine 3 (tRNALys3) bound to an mRNA with an AAA-codon in the A-site and Paromomycin | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
1EGA
 
 | | CRYSTAL STRUCTURE OF A WIDELY CONSERVED GTPASE ERA | | Descriptor: | PROTEIN (GTP-BINDING PROTEIN ERA), SULFATE ION | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1998-12-01 | | Release date: | 1999-07-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1FJG
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Wimberly, B.T, Morgan-Warren, R.J, Ramakrishnan, V. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics
Nature, 407, 2000
|
|
6FEC
 
 | | Human cap-dependent 48S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schaffitzel, C, Schaffitzel, C. | | Deposit date: | 2017-12-31 | | Release date: | 2018-03-14 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structure of a human cap-dependent 48S translation pre-initiation complex.
Nucleic Acids Res., 46, 2018
|
|
6FKR
 
 | | Crystal structure of the dolphin proline-rich antimicrobial peptide Tur1A bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16 ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Perebaskine, N, Benincasa, M, Gambato, S, Hofmann, S, Huter, P, Muller, C, Hilpert, K, Innis, C.A, Tossi, A, Wilson, D.N. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Dolphin Proline-Rich Antimicrobial Peptide Tur1A Inhibits Protein Synthesis by Targeting the Bacterial Ribosome.
Cell Chem Biol, 25, 2018
|
|
3OTO
 
 | | Crystal Structure of the 30S ribosomal subunit from a KsgA mutant of Thermus thermophilus (HB8) | | Descriptor: | 16S rRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Demirci, H, Murphy IV, F, Belardinelli, R, Kelley, A.C, Ramakrishnan, V, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2010-09-13 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function.
Rna, 16, 2010
|
|
6G5I
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State R | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
2XR1
 
 | | DIMERIC ARCHAEAL CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR WITH N-TERMINAL KH DOMAINS (KH-CPSF) FROM METHANOSARCINA MAZEI | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 100 KD SUBUNIT, ZINC ION | | Authors: | Mir-Montazeri, B, Ammelburg, M, Forouzan, D, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Dimeric Archaeal Cleavage and Polyadenylation Specificity Factor.
J.Struct.Biol., 173, 2011
|
|
2ZM6
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kaminishi, T, Wang, H, Kawazoe, M, Ishii, R, Schluenzen, F, Hanawa-Suetsugu, K, Wilson, D.N, Nomura, M, Takemoto, C, Shirouzu, M, Fucini, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-11 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the Thermus thermophilus 30S ribosomal subunit
To be Published
|
|
3R9X
 
 | | Crystal structure of Era in complex with MgGDPNP, nucleotides 1506-1542 of 16S ribosomal RNA, and KsgA | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, GTPase Era, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2011-03-26 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3' end of 16S rRNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2E5L
 
 | | A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine- Dalgarno interaction | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kaminishi, T, Wilson, D.N, Takemoto, C, Harms, J.M, Kawazoe, M, Schluenzen, F, Hanawa-Suetsugu, K, Shirouzu, M, Fucini, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-21 | | Release date: | 2007-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction
Structure, 15, 2007
|
|
2YKR
 
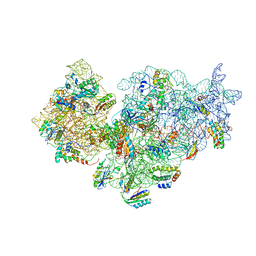 | | 30S ribosomal subunit with RsgA bound in the presence of GMPPNP | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Guo, Q, Yuan, Y, Xu, Y, Feng, B, Liu, L, Chen, K, Lei, J, Gao, N. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural Basis for the Function of a Small Gtpase Rsga on the 30S Ribosomal Subunit Maturation Revealed by Cryoelectron Microscopy.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3R9W
 
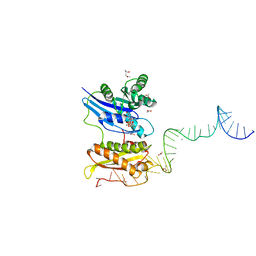 | |
9BDP
 
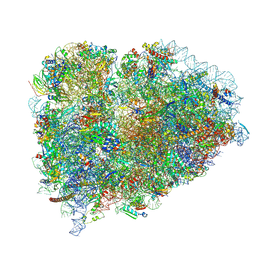 | |
8Y39
 
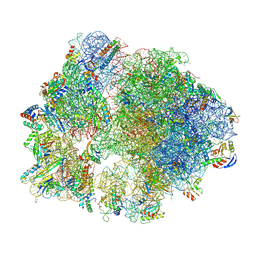 | | cryo-EM structure of Staphylococcus aureus(ATCC 29213) 70S ribosome in complex with MCX-190. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
9BDL
 
 | | 80S ribosome with angiogenin | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A. | | Deposit date: | 2024-04-12 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural mechanism of angiogenin activation by the ribosome.
Nature, 630, 2024
|
|
9FBV
 
 | |
