1VAZ
 
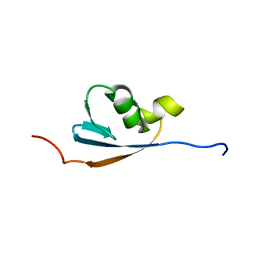 | | Solution structures of the p47 SEP domain | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-02-20 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97.
Embo J., 23, 2004
|
|
1VB0
 
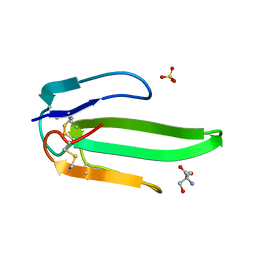 | | Atomic resolution structure of atratoxin-b, one short-chain neurotoxin from Naja atra | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cobrotoxin b, SULFATE ION | | Authors: | Lou, X, Liu, Q, Teng, M, Niu, L, Huang, Q, Hao, Q. | | Deposit date: | 2004-02-20 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
J.Biol.Chem., 279, 2004
|
|
1VB2
 
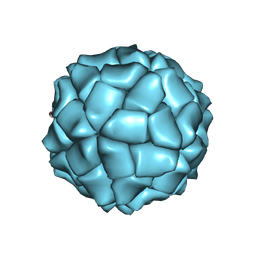 | | T=1 capsid structure of Sesbania mosaic virus coat protein deletion mutant CP-N(delta)65-D146N-D149N | | Descriptor: | coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumar, P.S, Vijay, C.S, Saravanan, V, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2004-02-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | T=1 capsid structures of Sesbania mosaic virus coat protein mutants: determinants of T=3 and T=1 capsid assembly
J.Mol.Biol., 342, 2004
|
|
1VB3
 
 | |
1VB4
 
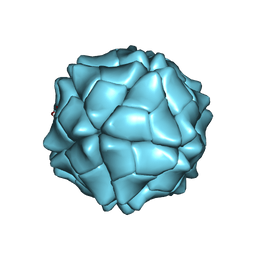 | | T=1 capsid structure of Sesbania mosaic virus coat protein deletion mutant CP-N(delta)36 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumar, P.S, Vijay, C.S, Saravanan, V, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2004-02-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | T=1 capsid structures of Sesbania mosaic virus coat protein mutants: determinants of T=3 and T=1 capsid assembly
J.Mol.Biol., 342, 2004
|
|
1VB5
 
 | |
1VB6
 
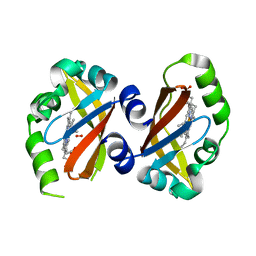 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (oxygen-bound form) | | Descriptor: | Heme pas sensor protein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Watanabe, M, Sagami, I, Mikami, B, Shimizu, T. | | Deposit date: | 2004-02-24 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of oxygen-bound form of a Heme PAS domain of Ec DOS
To be Published
|
|
1VB7
 
 | |
1VB8
 
 | |
1VB9
 
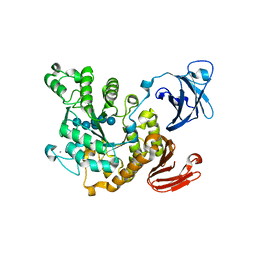 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) complexed with transglycosylated product | | Descriptor: | CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-[alpha-D-glucopyranose-(1-6)]alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-amylase II | | Authors: | Mizuno, M, Tonozuka, T, Uechi, A, Ohtaki, A, Ichikawa, K, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2004-02-25 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) complexed with transglycosylated product
EUR.J.BIOCHEM., 271, 2004
|
|
1VBA
 
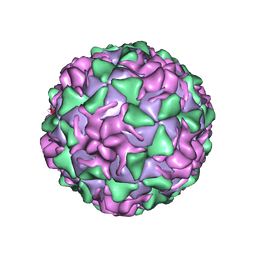 | | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R78206 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE PROPYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1VBB
 
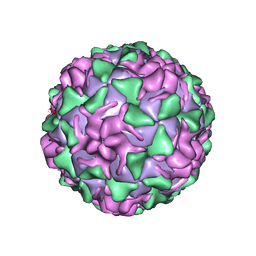 | | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R80633 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE BUTYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1VBC
 
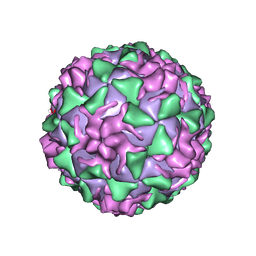 | | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R77975 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE ETHYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1VBD
 
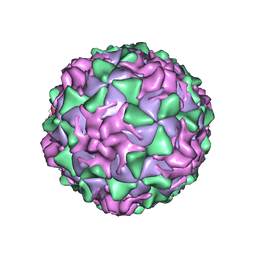 | | POLIOVIRUS (TYPE 1, MAHONEY STRAIN) COMPLEXED WITH R78206 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE PROPYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1VBE
 
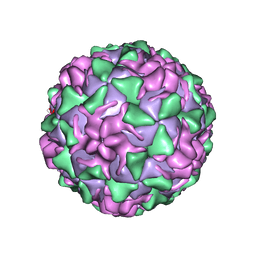 | | POLIOVIRUS (TYPE 3, SABIN STRAIN, MUTANT 242-H2) COMPLEXED WITH R78206 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE PROPYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1VBF
 
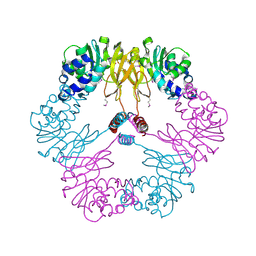 | | Crystal structure of protein L-isoaspartate O-methyltransferase homologue from Sulfolobus tokodaii | | Descriptor: | 231aa long hypothetical protein-L-isoaspartate O-methyltransferase | | Authors: | Tanaka, Y, Tsumoto, K, Yasutake, Y, Umetsu, M, Yao, M, Tanaka, I, Fukada, H, Kumagai, I. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How Oligomerization Contributes to the Thermostability of an Archaeon Protein: PROTEIN L-ISOASPARTYL-O-METHYLTRANSFERASE FROM SULFOLOBUS TOKODAII
J.Biol.Chem., 279, 2004
|
|
1VBG
 
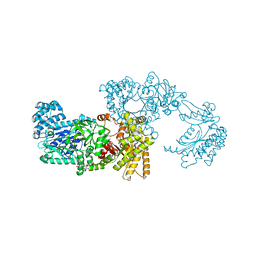 | | Pyruvate Phosphate Dikinase from Maize | | Descriptor: | MAGNESIUM ION, SULFATE ION, pyruvate,orthophosphate dikinase | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1VBH
 
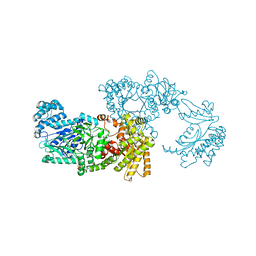 | | Pyruvate Phosphate Dikinase with bound Mg-PEP from Maize | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, SULFATE ION, ... | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1VBI
 
 | |
1VBJ
 
 | |
1VBK
 
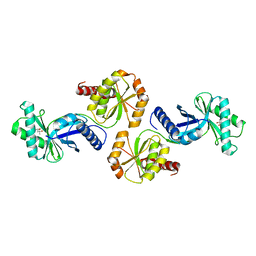 | | Crystal structure of PH1313 from Pyrococcus horikoshii Ot3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, hypothetical protein PH1313 | | Authors: | Sugahara, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-27 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Purification, crystallization and preliminary crystallographic analysis of the putative thiamine-biosynthesis protein PH1313 from Pyrococcus horikoshii OT3
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1VBL
 
 | | Structure of the thermostable pectate lyase PL 47 | | Descriptor: | CALCIUM ION, pectate lyase 47 | | Authors: | Nakaniwa, T, Tada, T, Yamaguchi, A, Kitatani, T, Takao, M, Sakai, T, Nishimura, K. | | Deposit date: | 2004-02-27 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Thermostability of Pectate Lyase from Bacillus sp. TS 47
To be Published
|
|
1VBM
 
 | | Crystal structure of the Escherichia coli tyrosyl-tRNA synthetase complexed with Tyr-AMS | | Descriptor: | 5'-O-[N-(L-TYROSYL)SULFAMOYL]ADENOSINE, SULFATE ION, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Sakamoto, K, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-27 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural snapshots of the KMSKS loop rearrangement for amino acid activation by bacterial tyrosyl-tRNA synthetase.
J.Mol.Biol., 346, 2005
|
|
1VBN
 
 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with Tyr-AMS | | Descriptor: | 5'-O-[N-(L-TYROSYL)SULFAMOYL]ADENOSINE, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Sakamoto, K, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-27 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1VBO
 
 | | Crystal structure of artocarpin-mannotriose complex | | Descriptor: | alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, artocarpin | | Authors: | Jeyaprakash, A.A, Srivastav, A, Surolia, A, Vijayan, M. | | Deposit date: | 2004-02-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity
J.Mol.Biol., 338, 2004
|
|
