5XZB
 
 | | Mouse cGAS bound to the inhibitor RU365 | | Descriptor: | (3R)-3-[1-(1H-benzimidazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
7RTO
 
 | | Cryo-EM structure of bluetongue virus capsid protein VP5 at low endosomal pH intermediate state 2 | | Descriptor: | Outer capsid protein VP5 | | Authors: | Xia, X, Wu, W.N, Cui, Y.X, Roy, P, Zhou, Z.H. | | Deposit date: | 2021-08-13 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Bluetongue virus capsid protein VP5 perforates membranes at low endosomal pH during viral entry.
Nat Microbiol, 6, 2021
|
|
1BQG
 
 | | THE STRUCTURE OF THE D-GLUCARATE DEHYDRATASE PROTEIN FROM PSEUDOMONAS PUTIDA | | Descriptor: | D-GLUCARATE DEHYDRATASE | | Authors: | Gulick, A.M, Palmer, D.R.J, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 1998-08-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structure of (D)-glucarate dehydratase from Pseudomonas putida.
Biochemistry, 37, 1998
|
|
5Z8S
 
 | |
5Z91
 
 | | Human mitochondrial ferritin mutant bound with gold ions | | Descriptor: | Ferritin, mitochondrial, GOLD ION, ... | | Authors: | Zang, J, Zheng, B, Zhao, G. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Design and site-directed compartmentalization of gold nanoclusters within the intrasubunit interfaces of ferritin nanocage.
J Nanobiotechnology, 17, 2019
|
|
1R6W
 
 | | Crystal structure of the K133R mutant of o-Succinylbenzoate synthase (OSBS) from Escherichia coli. Complex with SHCHC | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, MAGNESIUM ION, o-Succinylbenzoate Synthase | | Authors: | Klenchin, V.A, Taylor Ringia, E.A, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-10-17 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural and Mutagenic Studies of the Mechanism of the Reaction Catalyzed by o-Succinylbenzoate Synthase from Escherichia coli
Biochemistry, 42, 2003
|
|
3I7D
 
 | |
7NUF
 
 | | Vaccinia virus protein 018 in complex with STAT1 | | Descriptor: | ACETYL GROUP, SULFATE ION, Signal transducer and activator of transcription 1-alpha/beta, ... | | Authors: | Pantelejevs, T, Talbot-Cooper, C, Smith, G.L, Hyvonen, M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.0004015 Å) | | Cite: | Poxviruses and paramyxoviruses use a conserved mechanism of STAT1 antagonism to inhibit interferon signaling.
Cell Host Microbe, 30, 2022
|
|
3GPN
 
 | | Structure of the non-trimeric form of the E113G PCNA mutant protein | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Gakhar, L, Ramaswamy, S, Washington, M.T. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A charged residue at the subunit interface of PCNA promotes trimer formation by destabilizing alternate subunit interactions.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5Z8J
 
 | |
5Z8U
 
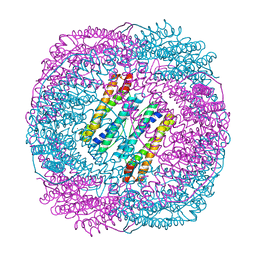 | | Human mitochondrial ferritin mutant - C102A/C130A | | Descriptor: | Ferritin, mitochondrial, MAGNESIUM ION | | Authors: | Zang, J, Zheng, B, Zhao, G. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and site-directed compartmentalization of gold nanoclusters within the intrasubunit interfaces of ferritin nanocage.
J Nanobiotechnology, 17, 2019
|
|
3GOH
 
 | |
3H2D
 
 | |
7NWJ
 
 | |
7NXP
 
 | |
3MFD
 
 | | The Structure of the Beta-lactamase superfamily domain of D-alanyl-D-alanine carboxypeptidase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, D-alanyl-D-alanine carboxypeptidase dacB | | Authors: | Cuff, M.E, Rakowski, E, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-01 | | Release date: | 2010-05-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the Beta-lactamase superfamily domain of D-alanyl-D-alanine carboxypeptidase from Bacillus subtilis.
TO BE PUBLISHED
|
|
5B5I
 
 | |
5IN9
 
 | | Crystal structure of Grp94 bound to methyl 3-chloro-2-(2-(1-((5-chlorofuran-2-yl)methyl)-1H-imidazol-2-yl)ethyl)-4,6-dihydroxybenzoate, an inhibitor based on the BnIm and Radamide scaffolds. | | Descriptor: | Endoplasmin, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Lieberman, R.L, Huard, D.J.E, Kizziah, J.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of Glucose Regulated Protein 94-Selective Inhibitors Based on the BnIm and Radamide Scaffold.
J.Med.Chem., 59, 2016
|
|
7C6C
 
 | | Crystal structure of native chitosanase from Bacillus subtilis MY002 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Chitosanase | | Authors: | Gou, Y, Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.258 Å) | | Cite: | Structure-based rational design of chitosanase CsnMY002 for high yields of chitobiose.
Colloids Surf B Biointerfaces, 202, 2021
|
|
7C6D
 
 | | Crystal structure of E19A mutant chitosanase from Bacillus subtilis MY002 complexed with 6 GlcN. | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Chitosanase | | Authors: | Gou, Y, Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structure-based rational design of chitosanase CsnMY002 for high yields of chitobiose.
Colloids Surf B Biointerfaces, 202, 2021
|
|
3NIO
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinobutyrase | | Descriptor: | Guanidinobutyrase, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
2HJP
 
 | |
2DUA
 
 | |
2HRW
 
 | | Crystal Structure of Phosphonopyruvate Hydrolase | | Descriptor: | CHLORIDE ION, Phosphonopyruvate hydrolase, SODIUM ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Kinetics of Phosphonopyruvate Hydrolase from Voriovorax sp. Pal2: New Insight into the Divergence of Catalysis within the PEP Mutase/Isocitrate Lyase Superfamily
Biochemistry, 45, 2006
|
|
6D6Z
 
 | |
