8G7L
 
 | | ATP-bound mtHsp60 V72I | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7O
 
 | | ATP- and mtHsp10-bound mtHsp60 V72I focus | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7N
 
 | | ATP- and mtHsp10-bound mtHsp60 V72I | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7J
 
 | | mtHsp60 V72I apo | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7K
 
 | | mtHsp60 V72I apo focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
6ZFD
 
 | | 14-3-3 zeta bound to the phosphorylated 18E6 C-terminus | | Descriptor: | 14-3-3 protein zeta/delta,Protein E6, GLYCEROL | | Authors: | Gogl, G, Tugaeva, K.V, Sluchanko, N.N, Trave, G. | | Deposit date: | 2020-06-17 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hierarchized phosphotarget binding by the seven human 14-3-3 isoforms.
Nat Commun, 12, 2021
|
|
6ZFG
 
 | | 14-3-3 zeta chimera with 18E6 and fusicoccin | | Descriptor: | 14-3-3 protein zeta/delta,Protein E6, FUSICOCCIN, GLYCEROL | | Authors: | Gogl, G, Tugaeva, K, Sluchanko, N.N, Trave, G. | | Deposit date: | 2020-06-17 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hierarchized phosphotarget binding by the seven human 14-3-3 isoforms.
Nat Commun, 12, 2021
|
|
8OLT
 
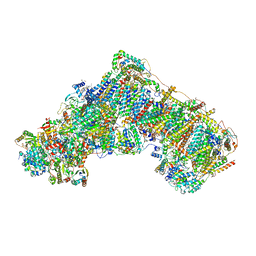 | | Mitochondrial complex I from Mus musculus in the active state bound with piericidin A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Grba, D.N, Chung, I, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-03-30 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Investigation of hydrated channels and proton pathways in a high-resolution cryo-EM structure of mammalian complex I.
Sci Adv, 9, 2023
|
|
8OM1
 
 | | Mitochondrial complex I from Mus musculus in the active state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Chung, I, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Investigation of hydrated channels and proton pathways in a high-resolution cryo-EM structure of mammalian complex I.
Sci Adv, 9, 2023
|
|
6RLZ
 
 | |
1I09
 
 | |
6ZU9
 
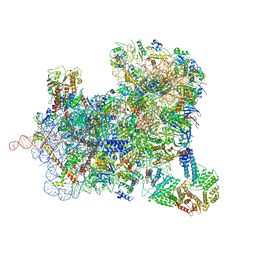 | | Structure of a yeast ABCE1-bound 48S initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
6EF5
 
 | | 14-3-3 with peptide | | Descriptor: | 14-3-3 protein zeta/delta, ARG-SER-LEU-SEP-ALA-PRO-GLY, LYS-LEU-SEP-LEU-GLN | | Authors: | Dhagat, U, Langendorf, C.G, Parker, M.W.P, Oakhill, J.S, Scott, J.W. | | Deposit date: | 2018-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | 14-3-3 with peptide
To Be Published
|
|
6EJL
 
 | |
5MPE
 
 | | 26S proteasome in presence of ATP (s2) | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN1, 26S proteasome regulatory subunit RPN10, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8J8Z
 
 | | Structure of beta-arrestin1 in complex with D6Rpp | | Descriptor: | Atypical chemokine receptor 2, Beta-arrestin-1, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
6RBE
 
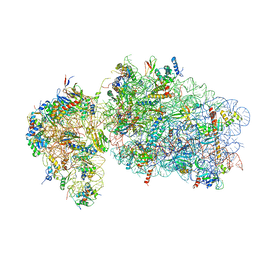 | | State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
1ADE
 
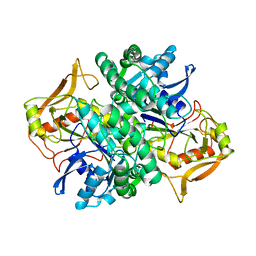 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE PH 7 AT 25 DEGREES CELSIUS | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE | | Authors: | Silva, M.M, Poland, B.W, Hoffman, C.M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1995-09-14 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structures of unligated adenylosuccinate synthetase from Escherichia coli.
J.Mol.Biol., 254, 1995
|
|
1L3R
 
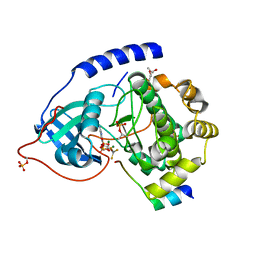 | | Crystal Structure of a Transition State Mimic of the Catalytic Subunit of cAMP-dependent Protein Kinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Madhusudan, Akamine, P, Xuong, N.-H, Taylor, S.S. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase.
Nat.Struct.Biol., 9, 2002
|
|
5MPC
 
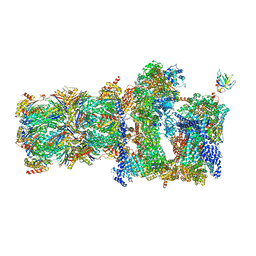 | | 26S proteasome in presence of BeFx (s4) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6EWW
 
 | |
6F08
 
 | | 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1) | | Descriptor: | 14-3-3 protein zeta/delta, PENTAETHYLENE GLYCOL, Son of sevenless homolog 1 | | Authors: | Ballone, A, Centorrino, F, Ottmann, C, Guo, S, Leysen, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1).
J. Struct. Biol., 202, 2018
|
|
6ZCE
 
 | | Structure of a yeast ABCE1-bound 43S pre-initiation complex | | Descriptor: | 18S ribosomal RNA (1719-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-07 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
1IB1
 
 | | CRYSTAL STRUCTURE OF THE 14-3-3 ZETA:SEROTONIN N-ACETYLTRANSFERASE COMPLEX | | Descriptor: | 14-3-3 ZETA ISOFORM, COA-S-ACETYL TRYPTAMINE, SEROTONIN N-ACETYLTRANSFERASE | | Authors: | Obsil, T, Ghirlando, R, Klein, D.C, Ganguly, S, Dyda, F. | | Deposit date: | 2001-03-26 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation.
Cell(Cambridge,Mass.), 105, 2001
|
|
5MPD
 
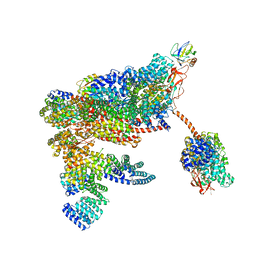 | | 26S proteasome in presence of ATP (s1) | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN1, 26S proteasome regulatory subunit RPN10, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
