1XAB
 
 | |
1UYQ
 
 | |
1XAA
 
 | |
1XGM
 
 | |
1T87
 
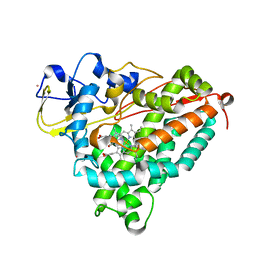 | | Crystal Structure of the Ferrous CO-bound Cytochrome P450cam (C334A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CARBON MONOXIDE, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1XRA
 
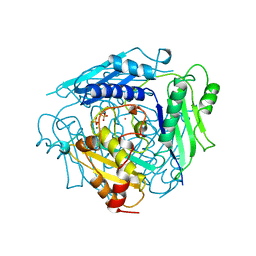 | | CRYSTAL STRUCTURE OF S-ADENOSYLMETHIONINE SYNTHETASE | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
1T8R
 
 | |
1XXC
 
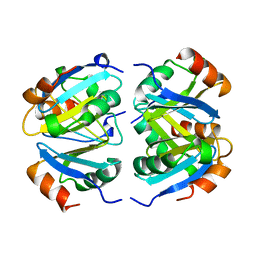 | |
1SZ9
 
 | |
1XPT
 
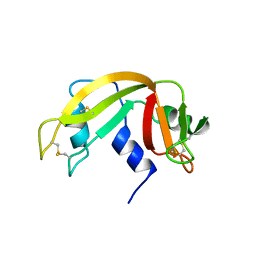 | | BOVINE RIBONUCLEASE A (PHOSPHATE-FREE) | | Descriptor: | RIBONUCLEASE A | | Authors: | Sadasivan, C, Nagendra, H.G, Vijayan, M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plasticity, hydration and accessibility in ribonuclease A. The structure of a new crystal form and its low-humidity variant.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1XUK
 
 | | TRYPSIN-BABIM-SULFATE, PH 5.9 | | Descriptor: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANE, CALCIUM ION, SULFATE ION, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-11-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
1XUG
 
 | | TRYPSIN-BABIM-ZN+2, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANE, CALCIUM ION, TRYPSIN, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
2EFK
 
 | | Crystal structure of the EFC domain of Cdc42-interacting protein 4 | | Descriptor: | Cdc42-interacting protein 4 | | Authors: | Shimada, A, Niwa, H, Chen, L, Liu, Z.-J, Wang, B.-C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis
Cell(Cambridge,Mass.), 129, 2007
|
|
1TAD
 
 | | GTPASE MECHANISM OF GPROTEINS FROM THE 1.7-ANGSTROM CRYSTAL STRUCTURE OF TRANSDUCIN ALPHA-GDP-ALF4- | | Descriptor: | CACODYLATE ION, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sondek, J, Lambright, D.G, Noel, J.P, Hamm, H.E, Sigler, P.B. | | Deposit date: | 1995-01-05 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4.
Nature, 372, 1994
|
|
1XRB
 
 | | S-adenosylmethionine synthetase (MAT, ATP: L-methionine S-adenosyltransferase, E.C.2.5.1.6) in which MET residues are replaced with selenomethionine residues (MSE) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
2EEU
 
 | | Guanine riboswitch U22A, A52U mutant bound to hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), Guanine riboswitch, ... | | Authors: | Gilbert, S.D, Love, C.E, Batey, R.T. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational analysis of the purine riboswitch aptamer domain
Biochemistry, 46, 2007
|
|
1TDY
 
 | |
1XPS
 
 | |
1TE5
 
 | |
1YEF
 
 | | STRUCTURE OF IGG2A FAB FRAGMENT (D2.3) COMPLEXED WITH SUBSTRATE ANALOGUE | | Descriptor: | IGG2A FAB FRAGMENT, PARA-NITROBENZYL GLUTARYL GLYCINIC ACID, ZINC ION | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 1997-05-29 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of a hydrolytic antibody and of complexes elucidate catalytic pathway from substrate binding and transition state stabilization through water attack and product release.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2EGW
 
 | |
1T0V
 
 | | NMR Solution Structure of the Engineered Lipocalin FluA(R95K) Northeast Structural Genomics Target OR17 | | Descriptor: | BILIN-BINDING PROTEIN | | Authors: | Mills, J.L, Liu, G, Skerra, A, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-04-13 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the engineered fluorescein-binding lipocalin FluA reveal rigidification of beta-barrel and variable loops upon enthalpy-driven ligand binding.
Biochemistry, 48, 2009
|
|
2DXV
 
 | |
1T8G
 
 | | Crystal structure of phage T4 lysozyme mutant L32A/L33A/T34A/C54T/C97A/E108V | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | He, M.M, Wood, Z.A, Baase, W.A, Xiao, H, Matthews, B.W. | | Deposit date: | 2004-05-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation.
Protein Sci., 13, 2004
|
|
1T8Y
 
 | |
