5TM5
 
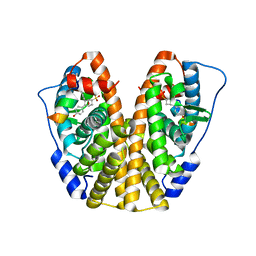 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 5-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)pentanoic acid | | Descriptor: | 5-{4-[(1S,4S,5R)-5-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}pentanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMQ
 
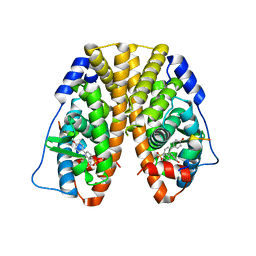 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Arene Core OBHS derivative, 4-bromophenyl 4,4''-dihydroxy-[1,1':2',1''-terphenyl]-4'-sulfonate | | Descriptor: | 4-bromophenyl 4,4''-dihydroxy-[1,1':2',1''-terphenyl]-4'-sulfonate, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TLX
 
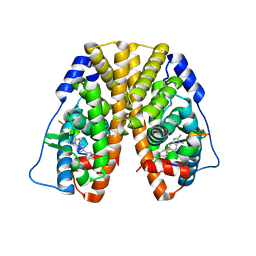 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 3,4-bis(4-hydroxyphenyl)thiophene 1,1-dioxide | | Descriptor: | 3,4-bis(4-hydroxyphenyl)-2,5-dihydro-1H-1lambda~6~-thiophene-1,1-dione, 3-methyl-6-phenyl-3H-imidazo[4,5-b]pyridin-2-amine, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM6
 
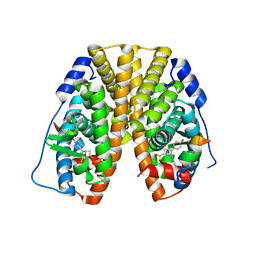 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 6-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)hexanoic acid | | Descriptor: | 6-{4-[(1S,4S,6R)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}hexanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-((1,3-dihydro-2H-inden-2-ylidene)methylene)diphenol | | Descriptor: | 4,4'-[(1,3-dihydro-2H-inden-2-ylidene)methylene]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN6
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Spiro BC-estradiol, (1S,1'S,3a'S,7a'S)-7a'-methyl-1',2,2',3,3',3a',4',6',7',7a'-decahydro-1,5'-spirobi[indene]-1',5-diol | | Descriptor: | (1S,1'S,3a'S,7a'S)-7a'-methyl-1',2,2',3,3',3a',4',6',7',7a'-decahydro-1,5'-spirobi[indene]-1',5-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
2NQ3
 
 | | Crystal structure of the C2 Domain of Human Itchy Homolog E3 Ubiquitin Protein Ligase | | Descriptor: | CHLORIDE ION, Itchy homolog E3 ubiquitin protein ligase | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The C2 Domain of Human Itchy Homolog E3 Ubiquitin Protein Ligase
To be Published
|
|
4W9D
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methyloxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4CI3
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4W9C
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(oxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 2) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
1IE9
 
 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to MC1288 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4W9H
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-acetamido-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 7) | | Descriptor: | N-acetyl-3-methyl-L-valyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4W9E
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(thiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 4) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
1M5B
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
9FTG
 
 | | Closed conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10 mM Ca2+ | | Descriptor: | CALCIUM ION, Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9FTH
 
 | | Open conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM Ca2+ | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9FTI
 
 | | Open conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM EDTA | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9FTJ
 
 | | Closed and disordered conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM EDTA | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
4CI1
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to thalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Thalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4W9L
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-((S)-2-acetamido-3,3-dimethylbutanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 15) | | Descriptor: | N-acetyl-3-methyl-L-valyl-3-methyl-L-valyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
8BD9
 
 | | Crystal structure of TRIM33 alpha PHD-Bromo domain in complex with 10 | | Descriptor: | 1,3-dimethylbenzimidazol-2-one, CALCIUM ION, E3 ubiquitin-protein ligase TRIM33, ... | | Authors: | Tassone, G, Pozzi, C, Palomba, T. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exploiting ELIOT for Scaffold-Repurposing Opportunities: TRIM33 a Possible Novel E3 Ligase to Expand the Toolbox for PROTAC Design.
Int J Mol Sci, 23, 2022
|
|
8BDY
 
 | | Crystal structure of TRIM33 alpha PHD-Bromo domain in complex with 9 | | Descriptor: | 1,3-dimethylbenzimidazol-2-one, CALCIUM ION, E3 ubiquitin-protein ligase TRIM33, ... | | Authors: | Tassone, G, Pozzi, C, Palomba, T. | | Deposit date: | 2022-10-20 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Exploiting ELIOT for Scaffold-Repurposing Opportunities: TRIM33 a Possible Novel E3 Ligase to Expand the Toolbox for PROTAC Design.
Int J Mol Sci, 23, 2022
|
|
8BD8
 
 | | Crystal structure of TRIM33 alpha PHD-Bromo domain in complex with 8 | | Descriptor: | 1,3-dimethylbenzimidazol-2-one, CALCIUM ION, E3 ubiquitin-protein ligase TRIM33, ... | | Authors: | Tassone, G, Pozzi, C, Palomba, T. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Exploiting ELIOT for Scaffold-Repurposing Opportunities: TRIM33 a Possible Novel E3 Ligase to Expand the Toolbox for PROTAC Design.
Int J Mol Sci, 23, 2022
|
|
4ZIP
 
 | | HIV-1 wild Type protease with GRL-0648A (a isophthalamide-derived P2-Ligand) | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[(2,5-dimethyl-1,3-oxazol-4-yl)methyl]-N'-[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]-N,5-dimethylbenzene-1,3-dicarboxamide, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure-based design, synthesis, X-ray studies, and biological evaluation of novel HIV-1 protease inhibitors containing isophthalamide-derived P2-ligands.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1BTJ
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE, APO FORM, CRYSTAL FORM 2 | | Descriptor: | PROTEIN (SERUM TRANSFERRIN) | | Authors: | Jeffrey, P.D, Bewley, M.C, Macgillivray, R.T.A, Mason, A.B, Woodworth, R.C, Baker, E.N. | | Deposit date: | 1998-09-01 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ligand-induced conformational change in transferrins: crystal structure of the open form of the N-terminal half-molecule of human transferrin.
Biochemistry, 37, 1998
|
|
