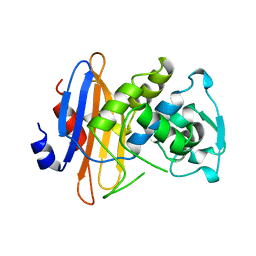
1BLP
 
 | |
140D
 
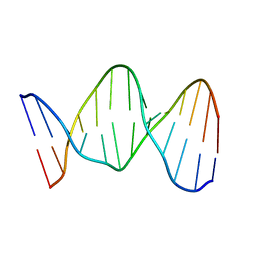 | | SOLUTION STRUCTURE OF A CONSERVED DNA SEQUENCE FROM THE HIV-1 GENOME: RESTRAINED MOLECULAR DYNAMICS SIMULATION WITH DISTANCE AND TORSION ANGLE RESTRAINTS DERIVED FROM TWO-DIMENSIONAL NMR SPECTRA | | Descriptor: | DNA (5'-D(*AP*GP*CP*TP*TP*GP*CP*CP*TP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*AP*AP*GP*GP*CP*AP*AP*GP*CP*T)-3') | | Authors: | Mujeeb, A, Kerwin, S.M, Kenyon, G.L, James, T.L. | | Deposit date: | 1993-09-24 | | Release date: | 1994-04-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conserved DNA sequence from the HIV-1 genome: restrained molecular dynamics simulation with distance and torsion angle restraints derived from two-dimensional NMR spectra.
Biochemistry, 32, 1993
|
|
142D
 
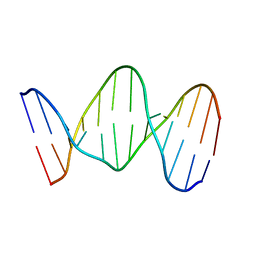 | |
141D
 
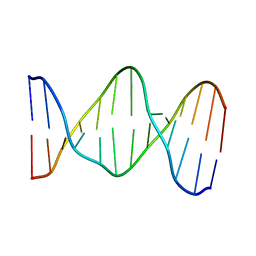 | |
1FRR
 
 | |
1FTA
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE, 1-PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH THE ALLOSTERIC INHIBITOR AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Lipscomb, W.N. | | Deposit date: | 1993-09-27 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric site of human liver fructose-1,6-bisphosphatase. Analysis of six AMP site mutants based on the crystal structure.
J.Biol.Chem., 269, 1994
|
|
1FRJ
 
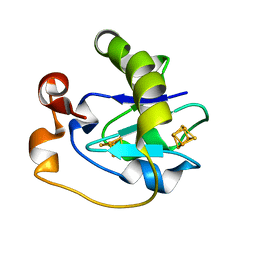 | |
1FRM
 
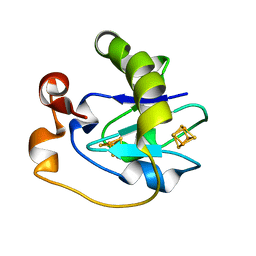 | |
1YMA
 
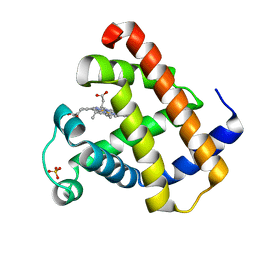 | |
1FRH
 
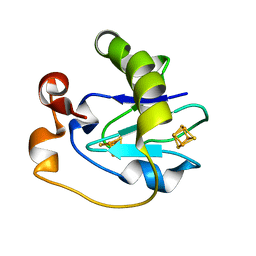 | |
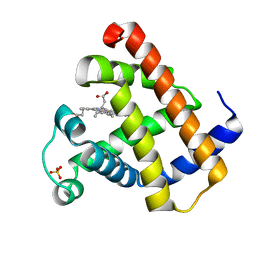
1YMB
 
 | |
1BLC
 
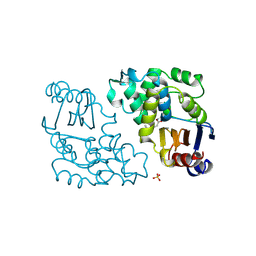 | |
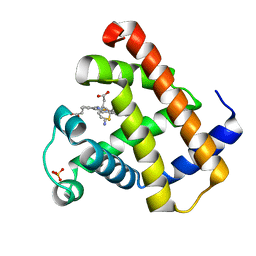
1YMC
 
 | |
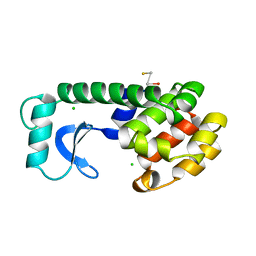
224L
 
 | |
1FRK
 
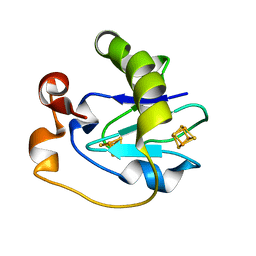 | |
1FRL
 
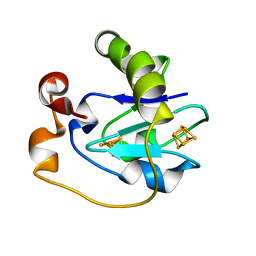 | |
1FRI
 
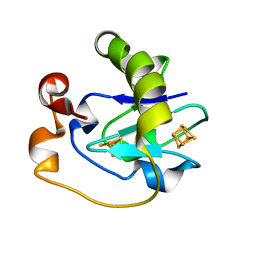 | |
139D
 
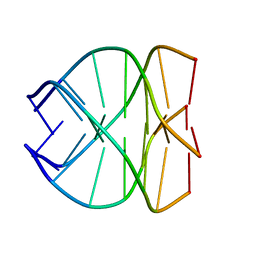 | |
2CYM
 
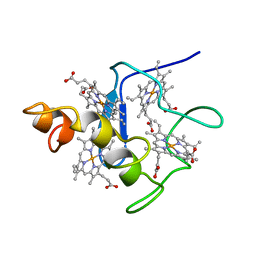 | | EFFECTS OF AMINO ACID SUBSTITUTION ON THREE-DIMENSIONAL STRUCTURE: AN X-RAY ANALYSIS OF CYTOCHROME C3 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH AT 2 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Morimoto, Y, Tani, T, Okumura, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1993-09-29 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of amino acid substitution on three-dimensional structure: an X-ray analysis of cytochrome c3 from Desulfovibrio vulgaris Hildenborough at 2 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
1ADN
 
 | |
1HPB
 
 | |
1BLH
 
 | |
1GOH
 
 | |
1DXI
 
 | | STRUCTURE DETERMINATION OF GLUCOSE ISOMERASE FROM STREPTOMYCES MURINUS AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION | | Authors: | Rasmussen, H, La Cour, T, Nyborg, J, Schulein, M. | | Deposit date: | 1993-09-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure determination of glucose isomerase from Streptomyces murinus at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1GOG
 
 | |
