8SWH
 
 | |
8SWX
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody Base4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Base4 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8SWW
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody IF3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NHP polyclonal antibody IF3 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8D4U
 
 | | Crystal Structure of Neutrophil Elastase Inhibited by Eap2 from S. aureus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Extracellular Adherence Protein, ... | | Authors: | Gido, C.D, Herdendorf, T.J, Geisbrecht, B.V. | | Deposit date: | 2022-06-02 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of two distinct neutrophil serine protease-binding modes within a Staphylococcus aureus innate immune evasion protein family.
J.Biol.Chem., 299, 2023
|
|
8SW4
 
 | | BG505 GT1.1 SOSIP in complex with NHP Fabs 21N13, 21M20 and RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 21M20 heavy chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Zhang, S, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Neutralizing antibodies induced in non-human primates by germline targeting Env trimer
To Be Published
|
|
6YC1
 
 | | Crystal structure of the H30A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
8D4Q
 
 | | Crystal Structure of Neutrophil Elastase Inhibited by Eap1 from S. aureus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Extracellular Adherence Protein, Neutrophil elastase | | Authors: | Gido, C.D, Herdendorf, T.J, Geisbrecht, B.V. | | Deposit date: | 2022-06-02 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of two distinct neutrophil serine protease-binding modes within a Staphylococcus aureus innate immune evasion protein family.
J.Biol.Chem., 299, 2023
|
|
1AW6
 
 | | GAL4 (CD), NMR, 24 STRUCTURES | | Descriptor: | CADMIUM ION, GAL4 (CD) | | Authors: | Baleja, J.D, Wagner, G. | | Deposit date: | 1997-10-10 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the DNA-binding domain of GAL4 and use of 3J(113Cd,1H) in structure determination.
J.Biomol.NMR, 10, 1997
|
|
8S0M
 
 | |
7OGU
 
 | | Plant peptide hormone receptor complex H1C9S1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roman, A.O, Jimenez-Sandoval, P, Santiago, J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | HSL1 and BAM1/2 impact epidermal cell development by sensing distinct signaling peptides.
Nat Commun, 13, 2022
|
|
8S2E
 
 | | Fab4251-DS-SOSIP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Nortier, P, Perez, L. | | Deposit date: | 2024-02-17 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | RAIN: a Machine Learning-based identification for HIV-1 bNAbs.
Res Sq, 2024
|
|
6SAU
 
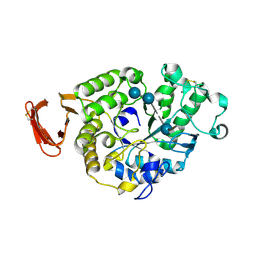 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, SODIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tinaqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
5IDL
 
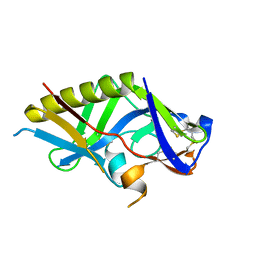 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Authors: | Julien, J.P, Ereno-Orbea, J, Jardine, J.G, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen.
Science, 351, 2016
|
|
1AOQ
 
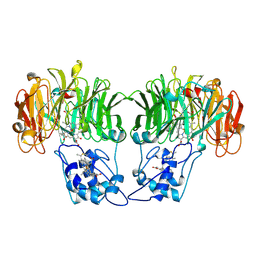 | |
1AOF
 
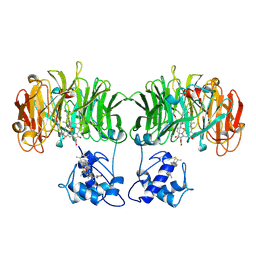 | | CYTOCHROME CD1 NITRITE REDUCTASE, REDUCED FORM | | Descriptor: | HEME D, NITRITE REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Williams, P.A, Fulop, V. | | Deposit date: | 1997-07-02 | | Release date: | 1997-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Haem-ligand switching during catalysis in crystals of a nitrogen-cycle enzyme.
Nature, 389, 1997
|
|
6YRB
 
 | | Crystal structure of the tetramerization domain of the glycoprotein Gn (Andes virus) at pH 7.5 | | Descriptor: | Envelope polyprotein, IODIDE ION, RNA (5'-D(*())-R(P*UP*UP*UP*())-3'), ... | | Authors: | Serris, A, Rey, F.A, Guardado-Calvo, P. | | Deposit date: | 2020-04-20 | | Release date: | 2020-10-14 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | The Hantavirus Surface Glycoprotein Lattice and Its Fusion Control Mechanism.
Cell, 183, 2020
|
|
1BET
 
 | | NEW PROTEIN FOLD REVEALED BY A 2.3 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF NERVE GROWTH FACTOR | | Descriptor: | BETA-NERVE GROWTH FACTOR | | Authors: | Mcdonald, N.Q, Lapatto, R, Murray-Rust, J, Gunning, J, Wlodawer, A, Blundell, T.L. | | Deposit date: | 1993-04-08 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor.
Nature, 354, 1991
|
|
1AOM
 
 | |
6YQ7
 
 | | Taka-amylase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
8SDH
 
 | |
5IBL
 
 | |
4X5C
 
 | | Anthranilate phosphoribosyltransferase variant R193L from Mycobacterium tuberculosis with pyrophosphate/PRPP and Mg2+ bound | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Cookson, T.V.M, Parker, E.J, Lott, J.S. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structures of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Variants Reveal the Conformational Changes That Facilitate Delivery of the Substrate to the Active Site.
Biochemistry, 54, 2015
|
|
7B2H
 
 | | Crystal structure of the methyl-coenzyme M reductase from Methanothermobacter Marburgensis derivatized with xenon | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-27 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
4X6P
 
 | |
8FOX
 
 | | AbeH (Tryptophan-5-halogenase) | | Descriptor: | SULFATE ION, Tryptophan 5-halogenase | | Authors: | Ashaduzzaman, M, Bellizzi, J.J. | | Deposit date: | 2023-01-03 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallographic and thermodynamic evidence of negative cooperativity of flavin and tryptophan binding in the flavin-dependent halogenases AbeH and BorH.
Biorxiv, 2023
|
|
