2BKJ
 
 | |
2BLH
 
 | | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
1YT4
 
 | | Crystal structure of TEM-76 beta-lactamase at 1.4 Angstrom resolution | | Descriptor: | Beta-lactamase TEM | | Authors: | Thomas, V.L, Golemi-Kotra, D, Kim, C, Vakulenko, S.B, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2005-02-09 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Consequences of the Inhibitor-Resistant Ser130Gly Substitution in TEM beta-Lactamase.
Biochemistry, 44, 2005
|
|
1YTC
 
 | |
1U5O
 
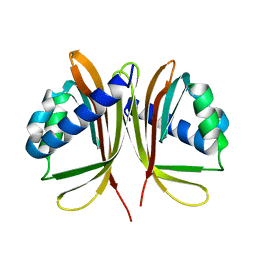 | | Structure of the D23A mutant of the nuclear transport carrier NTF2 | | Descriptor: | Nuclear transport factor 2 | | Authors: | Cushman, I, Bowman, B.R, Sowa, M.E, Lichtarge, O, Quiocho, F.A, Moore, M.S. | | Deposit date: | 2004-07-28 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational and biochemical identification of a nuclear pore complex binding site on the nuclear transport carrier NTF2.
J.Mol.Biol., 344, 2004
|
|
2BJW
 
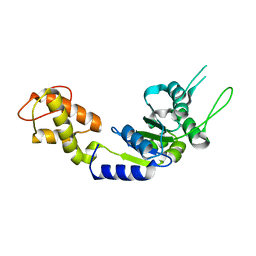 | | PspF AAA domain | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
2BND
 
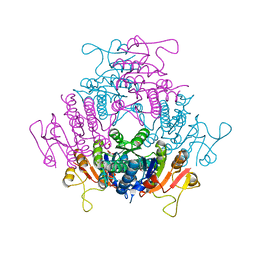 | | The structure of E. coli UMP kinase in complex with UDP | | Descriptor: | GLYCEROL, PYROPHOSPHATE 2-, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Briozzo, P, Evrin, C, Meyer, P, Assairi, L, Joly, N, Barzu, O, Gilles, A.M. | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia Coli Ump Kinase Differs from that of Other Nucleoside Monophosphate Kinases and Sheds New Light on Enzyme Regulation.
J.Biol.Chem., 280, 2005
|
|
1U65
 
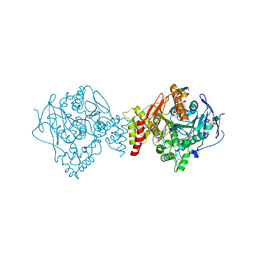 | | Ache W. CPT-11 | | Descriptor: | (4S)-4,11-DIETHYL-4-HYDROXY-3,14-DIOXO-3,4,12,14-TETRAHYDRO-1H-PYRANO[3',4':6,7]INDOLIZINO[1,2-B]QUINOLIN-9-YL 1,4'-BIPIPERIDINE-1'-CARBOXYLATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Harel, M, Hyatt, J.L, Brumshtein, B, Morton, C.L, Wadkins, R.W, Silman, I, Sussman, J.L, Potter, P.M, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-07-29 | | Release date: | 2005-07-19 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The crystal structure of the complex of the anticancer prodrug 7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothecin (CPT-11) with Torpedo californica acetylcholinesterase provides a molecular explanation for its cholinergic action
Mol.Pharmacol., 67, 2005
|
|
1YTN
 
 | | HYDROLASE | | Descriptor: | NITRATE ION, YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Authors: | Yuvaniyama, C, Fauman, E.B, Saper, M.A. | | Deposit date: | 1996-05-01 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray crystal structures of Yersinia tyrosine phosphatase with bound tungstate and nitrate. Mechanistic implications.
J.Biol.Chem., 271, 1996
|
|
2BXD
 
 | | Human serum albumin complexed with warfarin | | Descriptor: | R-WARFARIN, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
1U87
 
 | |
2C4N
 
 | | NagD from E.coli K-12 strain | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PROTEIN NAGD | | Authors: | Tremblay, L.W, Dunaway-Mariano, D, Allen, K. | | Deposit date: | 2005-10-20 | | Release date: | 2006-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Activity Analyses of Escherichia Coli K-12 Nagd Provide Insight Into the Evolution of Biochemical Function in the Haloalkanoic Acid Dehalogenase Superfamily
Biochemistry, 45, 2006
|
|
1YSD
 
 | | Yeast Cytosine Deaminase Double Mutant | | Descriptor: | CALCIUM ION, Cytosine deaminase, ZINC ION | | Authors: | Korkegian, A, Black, M.E, Baker, D, Stoddard, B.L. | | Deposit date: | 2005-02-08 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational thermostabilization of an enzyme.
Science, 308, 2005
|
|
2BHG
 
 | |
1U8M
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKYAS | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
2C62
 
 | |
1YTT
 
 | | YB SUBSTITUTED SUBTILISIN FRAGMENT OF MANNOSE BINDING PROTEIN-A (SUB-MBP-A), MAD STRUCTURE AT 110K | | Descriptor: | MANNOSE-BINDING PROTEIN A, YTTERBIUM (III) ION | | Authors: | Burling, F.T, Weis, W.I, Flaherty, K.M, Brunger, A.T. | | Deposit date: | 1995-11-09 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of protein solvation and discrete disorder with experimental crystallographic phases.
Science, 271, 1996
|
|
2C6D
 
 | | Aurora A kinase activated mutant (T287D) in complex with ADPNP | | Descriptor: | GLYCEROL, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pauptit, R.A, Pannifer, A.D, Breed, J, McMiken, H.H.J, Rowsell, S, Anderson, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-01-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sar and Inhibitor Complex Structure Determination of a Novel Class of Potent and Specific Aurora Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1UDV
 
 | | Crystal structure of the hyperthermophilic archaeal dna-binding protein Sso10b2 at 1.85 A | | Descriptor: | DNA binding protein SSO10b, ZINC ION | | Authors: | Chou, C.-C, Lin, T.-W, Chen, C.-Y, Wang, A.H.J. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the hyperthermophilic archaeal DNA-binding protein Sso10b2 at a resolution of 1.85 Angstroms
J.BACTERIOL., 185, 2003
|
|
1UAG
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE/:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J, Fanchon, E, Dideberg, O. | | Deposit date: | 1997-03-13 | | Release date: | 1998-03-18 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase from Escherichia coli.
EMBO J., 16, 1997
|
|
1YSL
 
 | | Crystal structure of HMG-CoA synthase from Enterococcus faecalis with AcetoAcetyl-CoA ligand. | | Descriptor: | ACETOACETIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Steussy, C.N, Vartia, A.A, Burgner II, J.W, Sutherlin, A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2005-02-08 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Crystal Structures of HMG-CoA Synthase from Enterococcus faecalis and a Complex with Its Second Substrate/Inhibitor Acetoacetyl-CoA.
Biochemistry, 44, 2005
|
|
2BUP
 
 | | T13G Mutant of the ATPASE fragment of Bovine HSC70 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sousa, M.C, Mckay, D.B. | | Deposit date: | 1998-09-08 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change.
Biochemistry, 37, 1998
|
|
1UEA
 
 | | MMP-3/TIMP-1 COMPLEX | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-3, TISSUE INHIBITOR OF METALLOPROTEINASE-1, ... | | Authors: | Bode, W, Maskos, K, Gomis-Rueth, F.-X, Nagase, H. | | Deposit date: | 1997-06-06 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1.
Nature, 389, 1997
|
|
2BVG
 
 | |
2BWC
 
 | |
