3GZI
 
 | |
3GY9
 
 | |
3GZ7
 
 | |
3H0N
 
 | |
4FMR
 
 | |
3GRD
 
 | |
3ISY
 
 | |
4GDZ
 
 | |
4GL3
 
 | |
4GBS
 
 | |
4GL6
 
 | |
3IIC
 
 | |
3G7P
 
 | |
4GCM
 
 | |
3GIW
 
 | |
3GB0
 
 | |
5HCQ
 
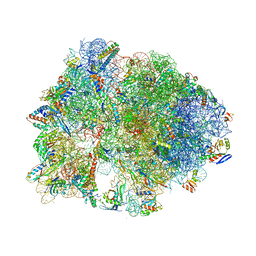 | | Crystal structure of antimicrobial peptide Oncocin d15-19 bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
3GN6
 
 | |
3G16
 
 | |
3G3L
 
 | |
3GFA
 
 | |
3G3S
 
 | |
3GGD
 
 | |
3G68
 
 | |
3G5B
 
 | | The structure of UNC5b cytoplasmic domain | | Descriptor: | Netrin receptor UNC5B, PHOSPHATE ION | | Authors: | Wang, R, Wei, Z, Zhang, M. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Autoinhibition of UNC5b revealed by the cytoplasmic domain structure of the receptor
Mol.Cell, 33, 2009
|
|
