1XVG
 
 | |
1TGX
 
 | | X-RAY STRUCTURE AT 1.55 A OF TOXIN GAMMA, A CARDIOTOXIN FROM NAJA NIGRICOLLIS VENOM. CRYSTAL PACKING REVEALS A MODEL FOR INSERTION INTO MEMBRANES | | Descriptor: | CHLORIDE ION, GAMMA-CARDIOTOXIN | | Authors: | Bilwes, A, Rees, B, Moras, D. | | Deposit date: | 1993-11-24 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure at 1.55 A of toxin gamma, a cardiotoxin from Naja nigricollis venom. Crystal packing reveals a model for insertion into membranes.
J.Mol.Biol., 239, 1994
|
|
2BST
 
 | | Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B2705 | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-27 ALPHA CHAIN PRECURSOR, ... | | Authors: | Stewart-Jones, G.B.E, diGleria, K, Kollnberger, S, McMichael, A.J, Bowness, P, Jones, E.Y. | | Deposit date: | 2005-05-23 | | Release date: | 2005-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures and Kir3Dl1 Recognition of Three Immunodominant Viral Peptides Complexed to Hla-B2705
Eur.J.Immunol., 35, 2005
|
|
1TFE
 
 | | DIMERIZATION DOMAIN OF EF-TS FROM T. THERMOPHILUS | | Descriptor: | ELONGATION FACTOR TS | | Authors: | Jiang, Y, Nock, S, Nesper, M, Sprinzl, M, Sigler, P.B. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and importance of the dimerization domain in elongation factor Ts from Thermus thermophilus.
Biochemistry, 35, 1996
|
|
1SZJ
 
 | |
2BTI
 
 | | Structure-function studies of the RmsA CsrA post-transcriptional global regulator protein family reveals a class of RNA-binding structure | | Descriptor: | ACETATE ION, CARBON STORAGE REGULATOR HOMOLOG, SULFATE ION | | Authors: | Heeb, S, Kuehne, S.A, Bycroft, M, Crivii, S, Allen, M.D, Haas, D, Camara, M, Williams, P. | | Deposit date: | 2005-05-31 | | Release date: | 2006-01-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Analysis of the Post-Transcriptional Regulator Rsma Reveals a Novel RNA-Binding Site.
J.Mol.Biol., 355, 2006
|
|
1TKE
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase complexed with serine | | Descriptor: | SERINE, Threonyl-tRNA synthetase | | Authors: | Dock-Bregeon, A.C, Rees, B, Torres-Larios, A, Bey, G, Caillet, J, Moras, D. | | Deposit date: | 2004-06-08 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Achieving Error-Free Translation; The Mechanism of Proofreading of Threonyl-tRNA Synthetase at Atomic Resolution.
Mol.Cell, 16, 2004
|
|
1X9K
 
 | | An all-RNA Hairpin Ribozyme with mutation U39C | | Descriptor: | 5'-R(*AP*AP*UP*AP*GP*AP*GP*AP*AP*GP*CP*GP*A)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*GP*CP*AP*GP*UP*CP*CP*UP*AP*UP*U)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2004-08-21 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Conformational Heterogeneity at Position U37 of an All-RNA Hairpin Ribozyme with Implications for Metal Binding and the Catalytic Structure of the S-Turn.
Biochemistry, 44, 2005
|
|
2C9L
 
 | | Structure of the Epstein-Barr virus ZEBRA protein | | Descriptor: | 5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*CP *TP*CP*AP*TP*GP*AP*AP*GP*T)-3', 5'-D(*AP*CP*TP*TP*CP*AP*CP*TP*GP*AP *GP*TP*CP*AP*GP*TP*GP*CP*T)-3', BZLF1 TRANS-ACTIVATOR PROTEIN | | Authors: | Petosa, C, Morand, P, Baudin, F, Moulin, M, Artero, J.B, Muller, C.W. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Lytic Cycle Activation by the Epstein-Barr Virus Zebra Protein
Mol.Cell, 21, 2006
|
|
1SRR
 
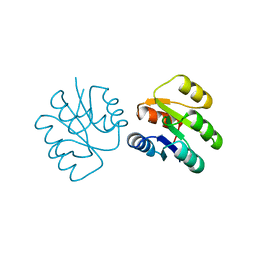 | | CRYSTAL STRUCTURE OF A PHOSPHATASE RESISTANT MUTANT OF SPORULATION RESPONSE REGULATOR SPO0F FROM BACILLUS SUBTILIS | | Descriptor: | CALCIUM ION, SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Whiteley, J.M, Hoch, J.A, Zapf, J, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1996-04-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis.
Structure, 4, 1996
|
|
2CAU
 
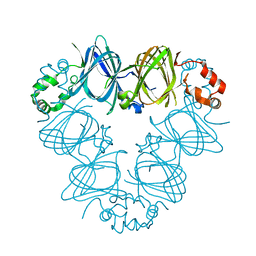 | | CANAVALIN FROM JACK BEAN | | Descriptor: | PROTEIN (CANAVALIN) | | Authors: | Ko, T.-P, Day, J, Macpherson, A. | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1XCA
 
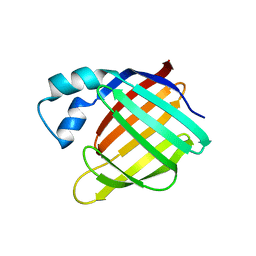 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN TYPE II | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1996-12-31 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of apo-cellular retinoic acid-binding protein type II (R111M) suggests a mechanism of ligand entry.
J.Mol.Biol., 278, 1998
|
|
1STN
 
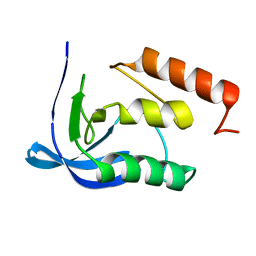 | |
2C7B
 
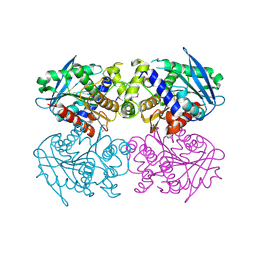 | | The Crystal Structure of EstE1, a New Thermophilic and Thermostable Carboxylesterase Cloned from a Metagenomic Library | | Descriptor: | CARBOXYLESTERASE | | Authors: | Byun, J.-S, Rhee, J.-K, Kim, D.-U, Oh, J.-W, Cho, H.-S. | | Deposit date: | 2005-11-21 | | Release date: | 2005-12-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Hyperthermophilic Esterase Este1 and the Relationship between its Dimerization and Thermostability Properties.
Bmc Struct.Biol., 7, 2007
|
|
2C8V
 
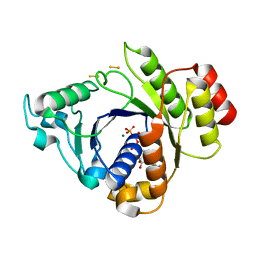 | | Insights into the role of nucleotide-dependent conformational change in nitrogenase catalysis: Structural characterization of the nitrogenase Fe protein Leu127 deletion variant with bound MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Sen, S, Krishnakumar, A, McClead, J, Johnson, M.K, Seefeldt, L.C, Szilagyi, R.K, Peters, J.W. | | Deposit date: | 2005-12-08 | | Release date: | 2006-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into the Role of Nucleotide-Dependent Conformational Change in Nitrogenase Catalysis: Structural Characterization of the Nitrogenase Fe Protein Leu127 Deletion Variant with Bound Mgatp.
J.Inorg.Biochem., 100, 2006
|
|
1XDQ
 
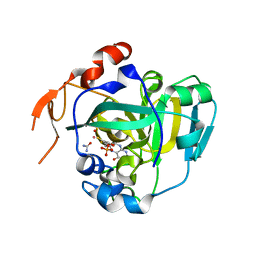 | | Structural and Biochemical Identification of a Novel Bacterial Oxidoreductase | | Descriptor: | Bacterial Sulfite Oxidase, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Loschi, L, Brokx, S.J, Hills, T.L, Zhang, G, Bertero, M.G, Lovering, A.L, Weiner, J.H, Strynadka, N.C. | | Deposit date: | 2004-09-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and biochemical identification of a novel bacterial oxidoreductase.
J.Biol.Chem., 279, 2004
|
|
2CGP
 
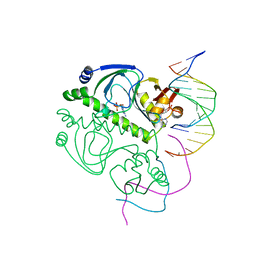 | | CATABOLITE GENE ACTIVATOR PROTEIN/DNA COMPLEX, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*AP*AP*TP*GP*TP*GP*AP*CP*AP*TP*AP*T)-3'), DNA (5'-D(*GP*TP*CP*AP*CP*AP*TP*TP*AP*AP*T)-3'), ... | | Authors: | Passner, J.M, Steitz, T.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1SUR
 
 | | PHOSPHO-ADENYLYL-SULFATE REDUCTASE | | Descriptor: | PAPS REDUCTASE | | Authors: | Sinning, I, Savage, H. | | Deposit date: | 1998-04-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphoadenylyl sulphate (PAPS) reductase: a new family of adenine nucleotide alpha hydrolases.
Structure, 5, 1997
|
|
2CFM
 
 | |
1XDM
 
 | | Structure of human aldolase B associated with hereditary fructose intolerance (A149P), at 291K | | Descriptor: | Fructose-bisphosphate aldolase B, SULFATE ION | | Authors: | Malay, A.D, Allen, K.N, Tolan, D.R. | | Deposit date: | 2004-09-07 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the thermolabile mutant aldolase B, A149P: molecular basis of hereditary fructose intolerance.
J.Mol.Biol., 347, 2005
|
|
2CJX
 
 | | Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis | | Descriptor: | CASPASE-3, PHQ-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE | | Authors: | Ganesan, R, Mittl, P.R.E, Jelakovic, S, Grutter, M.G. | | Deposit date: | 2006-04-09 | | Release date: | 2006-06-27 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extended Substrate Recognition in Caspase-3 Revealed by High Resolution X-Ray Structure Analysis
J.Mol.Biol., 359, 2006
|
|
2CM7
 
 | | Structural Basis for Inhibition of Protein Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics | | Descriptor: | ISOTHIAZOLIDINONE ANALOG, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
1SWV
 
 | | Crystal structure of the D12A mutant of phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
2CHB
 
 | | CHOLERA TOXIN B-PENTAMER COMPLEXED WITH GM1 PENTASACCHARIDE | | Descriptor: | CHOLERA TOXIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 1997-06-03 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of receptor binding by cholera toxin mutants.
Protein Sci., 6, 1997
|
|
1XUF
 
 | | TRYPSIN-BABIM-ZN+2, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANE ZINC, CALCIUM ION, TRYPSIN | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
