3RVK
 
 | | Structure of the CheY-Mn2+ Complex with substitutions at 59 and 89: N59D E89Q | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVP
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89K | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVS
 
 | | Structure of the CheYN59D/E89R Tungstate complex | | Descriptor: | Chemotaxis protein CheY, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVR
 
 | | Structure of the CheYN59D/E89R Molybdate complex | | Descriptor: | Chemotaxis protein CheY, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVL
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89R | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVJ
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89Q | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVO
 
 | | Structure of CheY-Mn2+ Complex with substitutions at 59 and 89: N59D E89Y | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVM
 
 | | Structure of the CheY-Mn2+ Complex with substitutions at 59 and 89: N59D and E89R | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVN
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89Y | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3T6K
 
 | |
3T8Y
 
 | |
3RVQ
 
 | | Structure of the CheY-Mn2+ Complex with substitutions at 59 and 89: N59D E89K | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3TMY
 
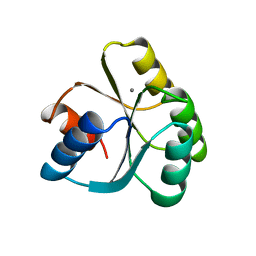 | | CHEY FROM THERMOTOGA MARITIMA (MN-III) | | Descriptor: | CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-04 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
3TO5
 
 | |
2IYN
 
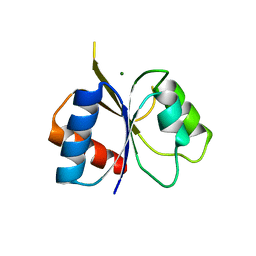 | | The co-factor-induced pre-active conformation in PhoB | | Descriptor: | MAGNESIUM ION, PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Sola, M, Drew, D.L, Blanco, A.G, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 2006-07-19 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Cofactor-Induced Pre-Active Conformation in Phob.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IDM
 
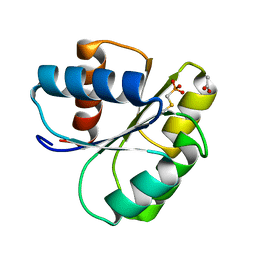 | | 2.00 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | ACETATE ION, Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2MSL
 
 | |
2ID9
 
 | | 1.85 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2N9U
 
 | |
2JVK
 
 | |
2JRL
 
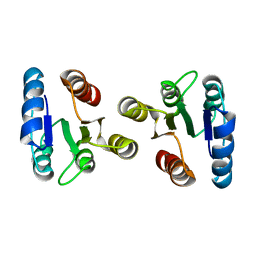 | | Solution structure of the beryllofluoride-activated NtrC4 receiver domain dimer | | Descriptor: | Transcriptional regulator (NtrC family) | | Authors: | Lee, C, Hong, E, Doucleff, M, Pelton, J.G, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Beryllofluoride-Activated NtrC4 Receiver Domain Dimer.
To be Published
|
|
2JVI
 
 | |
2MSK
 
 | |
2I6F
 
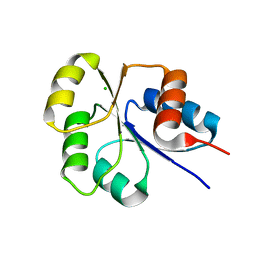 | | Receiver domain from Myxococcus xanthus social motility protein FrzS | | Descriptor: | CHLORIDE ION, Response regulator FrzS | | Authors: | Echols, N, Fraser, J, Merlie, J, Zusman, D, Alber, T. | | Deposit date: | 2006-08-28 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
2MSW
 
 | |
