1YPR
 
 | |
1YDV
 
 | | TRIOSEPHOSPHATE ISOMERASE (TIM) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Velankar, S.S, Murthy, M.R.N. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triosephosphate isomerase from Plasmodium falciparum: the crystal structure provides insights into antimalarial drug design.
Structure, 5, 1997
|
|
1YST
 
 | | STRUCTURE OF THE PHOTOCHEMICAL REACTION CENTER OF A SPHEROIDENE CONTAINING PURPLE BACTERIUM, RHODOBACTER SPHAEROIDES Y, AT 3 ANGSTROMS RESOLUTION | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, MANGANESE (II) ION, ... | | Authors: | Arnoux, B, Gaucher, J.F, Ducruix, A, Reiss-Husson, F. | | Deposit date: | 1994-12-07 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the photochemical reaction centre of a spheroidene-containing purple-bacterium, Rhodobacter sphaeroides Y, at 3 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1YET
 
 | |
1YPT
 
 | | CRYSTAL STRUCTURE OF YERSINIA PROTEIN TYROSINE PHOSPHATASE AT 2.5 ANGSTROMS AND THE COMPLEX WITH TUNGSTATE | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YERSINIA (CATALYTIC DOMAIN) | | Authors: | Stuckey, J.A, Schubert, H.L, Fauman, E.B, Zhang, Z.-Y, Dixon, J.E, Saper, M.A. | | Deposit date: | 1994-09-16 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate.
Nature, 370, 1994
|
|
1YGP
 
 | | PHOSPHORYLATED FORM OF YEAST GLYCOGEN PHOSPHORYLASE WITH PHOSPHATE BOUND IN THE ACTIVE SITE. | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, YEAST GLYCOGEN PHOSPHORYLASE | | Authors: | Lin, K, Rath, V.L, Dai, S.C, Fletterick, R.J, Hwang, P.K. | | Deposit date: | 1996-05-30 | | Release date: | 1996-12-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A protein phosphorylation switch at the conserved allosteric site in GP.
Science, 273, 1996
|
|
1ZCF
 
 | |
1YZ0
 
 | | R-State AMP Complex Reveals Initial Steps of the Quaternary Transition of Fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase, ... | | Authors: | Iancu, C.V, Mukund, S, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | R-state AMP complex reveals initial steps of the quaternary transition of fructose-1,6-bisphosphatase.
J.Biol.Chem., 280, 2005
|
|
1Z1W
 
 | | Crystal structures of the tricorn interacting facor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 394, 2005
|
|
1ZHV
 
 | | X-ray Crystal Structure Protein Atu0741 from Agobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR8. | | Descriptor: | hypothetical protein Atu0741 | | Authors: | Vorobiev, S.M, Kuzin, A, Abashidze, M, Forouhar, F, Xio, R, Ma, L.-C, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-26 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of hypothetical protein Atu071 from Agobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR8.
To be Published
|
|
1Z34
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1ZIY
 
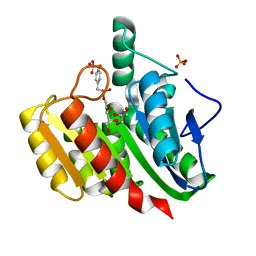 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (C123S) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.9 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2FC0
 
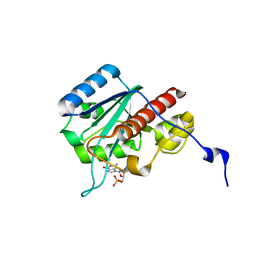 | | WRN exonuclease, Mn dGMP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Werner syndrome helicase | | Authors: | Perry, J.J, Tainer, J.A. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FBY
 
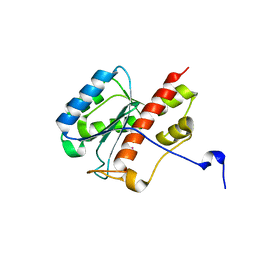 | | WRN exonuclease, Eu complex | | Descriptor: | EUROPIUM (III) ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FCZ
 
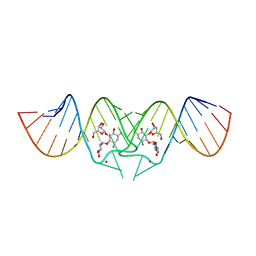 | | HIV-1 DIS kissing-loop in complex with ribostamycin | | Descriptor: | HIV-1 DIS RNA, POTASSIUM ION, RIBOSTAMYCIN | | Authors: | Ennifar, E, Paillart, J.C, Marquet, R, Dumas, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell.
Nucleic Acids Res., 34, 2006
|
|
1Z9L
 
 | | 1.7 Angstrom Crystal Structure of the Rat VAP-A MSP Homology Domain | | Descriptor: | Vesicle-associated membrane protein-associated protein A | | Authors: | Kaiser, S.E, Brickner, J.H, Reilein, A.R, Fenn, T.D, Walter, P, Brunger, A.T. | | Deposit date: | 2005-04-03 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of FFAT motif-mediated ER targeting
Structure, 13, 2005
|
|
1ZAP
 
 | | SECRETED ASPARTIC PROTEASE FROM C. ALBICANS | | Descriptor: | N-ethyl-N-[(4-methylpiperazin-1-yl)carbonyl]-D-phenylalanyl-N-[(1S,2S,4R)-4-(butylcarbamoyl)-1-(cyclohexylmethyl)-2-hydroxy-5-methylhexyl]-L-norleucinamide, SECRETED ASPARTIC PROTEINASE, ZINC ION | | Authors: | Abad-Zapatero, C, Muchmore, S.W. | | Deposit date: | 1996-01-16 | | Release date: | 1997-04-21 | | Last modified: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a secreted aspartic protease from C. albicans complexed with a potent inhibitor: implications for the design of antifungal agents.
Protein Sci., 5, 1996
|
|
1Z1F
 
 | | Crystal structure of stilbene synthase from Arachis hypogaea (resveratrol-bound form) | | Descriptor: | CITRIC ACID, RESVERATROL, stilbene synthase | | Authors: | Shomura, Y, Torayama, I, Suh, D.Y, Xiang, T, Kita, A, Sankawa, U, Miki, K. | | Deposit date: | 2005-03-03 | | Release date: | 2005-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of stilbene synthase from Arachis hypogaea
Proteins, 60, 2005
|
|
1Z33
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase | | Descriptor: | purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1YY3
 
 | |
1Z39
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2'-deoxyinosine | | Descriptor: | 2'-DEOXYINOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1YZ3
 
 | | Structure of human pnmt complexed with cofactor product adohcy and inhibitor SK&F 64139 | | Descriptor: | 7,8-DICHLORO-1,2,3,4-TETRAHYDROISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, Q, Gee, C.L, Lin, F, Martin, J.L, Grunewald, G.L, McLeish, M.J. | | Deposit date: | 2005-02-27 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, mutagenic, and kinetic analysis of the binding of substrates and inhibitors of human phenylethanolamine N-methyltransferase
J.Med.Chem., 48, 2005
|
|
1YZY
 
 | |
1Z8P
 
 | | Ferrous dioxygen complex of the A245S cytochrome P450eryF | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, 6-deoxyerythronolide B hydroxylase, OXYGEN MOLECULE, ... | | Authors: | Poulos, T.L, Nagano, S. | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the ferrous dioxygen complex of wild-type cytochrome P450eryF and its mutants, A245S and A245T: investigation of the proton transfer system in P450eryF.
J.Biol.Chem., 280, 2005
|
|
1ZBI
 
 | | Bacillus halodurans RNase H catalytic domain mutant D132N in complex with 12-mer RNA/DNA hybrid | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3', 5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3', MAGNESIUM ION, ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
