1ASP
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
6INS
 
 | | X-RAY ANALYSIS OF THE SINGLE CHAIN B29-A1 PEPTIDE-LINKED INSULIN MOLECULE. A COMPLETELY INACTIVE ANALOGUE | | Descriptor: | INSULIN, ZINC ION | | Authors: | Derewenda, U, Derewenda, Z, Dodson, E.J, Dodson, G.G, Bing, X, Markussen, J. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analysis of the single chain B29-A1 peptide-linked insulin molecule. A completely inactive analogue.
J.Mol.Biol., 220, 1991
|
|
1ASO
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
2PDE
 
 | | THE HIGH RESOLUTION STRUCTURE OF THE PERIPHERAL SUBUNIT-BINDING DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM THE PYRUVATE DEHYDROGENASE MULTIENZYME COMPLEX OF BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Kalia, Y.N, Brocklehurst, S.M, Hipps, D.S, Appella, E, Sakaguchi, K, Perham, R.N. | | Deposit date: | 1992-11-25 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The high-resolution structure of the peripheral subunit-binding domain of dihydrolipoamide acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus.
J.Mol.Biol., 230, 1993
|
|
1TPL
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF TYROSINE PHENOL-LYASE | | Descriptor: | SULFATE ION, TYROSINE PHENOL-LYASE | | Authors: | Antson, A, Demidkina, T, Dauter, Z, Harutyunyan, E, Wilson, K. | | Deposit date: | 1992-11-25 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of tyrosine phenol-lyase.
Biochemistry, 32, 1993
|
|
1ASQ
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, AZIDE ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
2PDD
 
 | | THE HIGH RESOLUTION STRUCTURE OF THE PERIPHERAL SUBUNIT-BINDING DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM THE PYRUVATE DEHYDROGENASE MULTIENZYME COMPLEX OF BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Kalia, Y.N, Brocklehurst, S.M, Hipps, D.S, Appella, E, Sakaguchi, K, Perham, R.N. | | Deposit date: | 1992-11-25 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The high-resolution structure of the peripheral subunit-binding domain of dihydrolipoamide acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus.
J.Mol.Biol., 230, 1993
|
|
1AEP
 
 | |
1ERP
 
 | | NUCLEAR MAGNETIC RESONANCE SOLUTION STRUCTURE OF THE PHEROMONE ER-10 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | Descriptor: | PHEROMONE ER-10 | | Authors: | Brown, L.R, Mronga, S, Bradshaw, R, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 1992-12-02 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the pheromone Er-10 from the ciliated protozoan Euplotes raikovi.
J.Mol.Biol., 231, 1993
|
|
1FKD
 
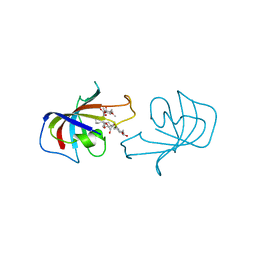 | | FK-506 BINDING PROTEIN: THREE-DIMENSIONAL STRUCTURE OF THE COMPLEX WITH THE ANTAGONIST L-685,818 | | Descriptor: | 18-HYDROXYASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Becker, J.W, Rotonda, J, Mckeever, B.M. | | Deposit date: | 1992-12-02 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | FK-506-binding protein: three-dimensional structure of the complex with the antagonist L-685,818.
J.Biol.Chem., 268, 1993
|
|
1MAT
 
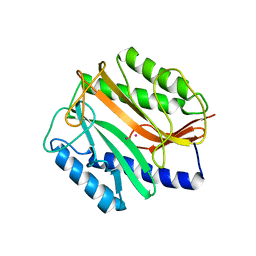 | |
1HVN
 
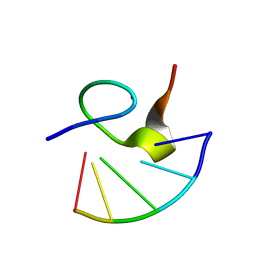 | |
1HVO
 
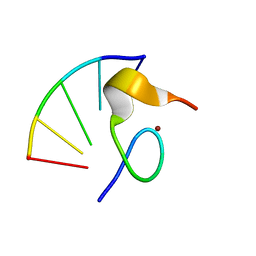 | |
1LGA
 
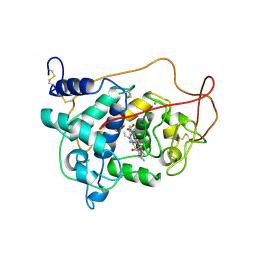 | | CRYSTALLOGRAPHIC REFINEMENT OF LIGNIN PEROXIDASE AT 2 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LIGNIN PEROXIDASE, ... | | Authors: | Poulos, T.L, Edwards, S.L, Wariishi, H, Gold, M.H. | | Deposit date: | 1992-12-08 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystallographic refinement of lignin peroxidase at 2 A.
J.Biol.Chem., 268, 1993
|
|
1OPA
 
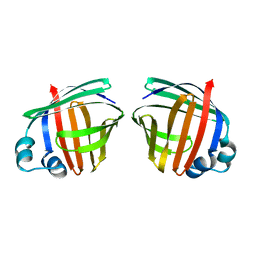 | |
1OPB
 
 | |
3INK
 
 | |
1NOA
 
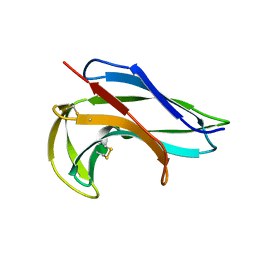 | |
1TCH
 
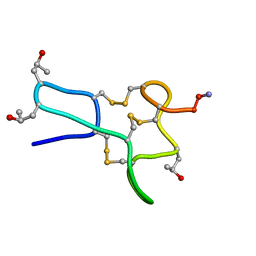 | |
1TCJ
 
 | |
1TCG
 
 | |
1TCK
 
 | |
1GRA
 
 | |
1GRH
 
 | | INHIBITION OF HUMAN GLUTATHIONE REDUCTASE BY THE NITROSOUREA DRUGS 1,3-BIS(2-CHLOROETHYL)-1-NITROSOUREA AND 1-(2-CHLOROETHYL)-3-(2-HYDROXYETHYL)-1-NITROSOUREA | | Descriptor: | ETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, ... | | Authors: | Karplus, P.A, Schulz, G.E. | | Deposit date: | 1992-12-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of human glutathione reductase by the nitrosourea drugs 1,3-bis(2-chloroethyl)-1-nitrosourea and 1-(2-chloroethyl)-3-(2-hydroxyethyl)-1-nitrosourea. A crystallographic analysis.
Eur.J.Biochem., 171, 1988
|
|
1GRF
 
 | |
