2N16
 
 | |
5LPX
 
 | | Crystal structure of PKC phosphorylation-mimicking mutant (S26E) Annexin A2 | | Descriptor: | Annexin A2, CALCIUM ION, GLYCEROL | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
5LM9
 
 | | Structure of E. coli NusA | | Descriptor: | MAGNESIUM ION, SULFATE ION, Transcription termination/antitermination protein NusA | | Authors: | Said, N, Weber, G, Santos, K, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
7N9Z
 
 | | E. coli cytochrome bo3 in MSP nanodisc | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vallese, F, Clarke, O.B. | | Deposit date: | 2021-06-19 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo 3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5LPU
 
 | | Crystal structure of Annexin A2 complexed with S100A4 | | Descriptor: | Annexin A2, CALCIUM ION, GLYCEROL, ... | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
7NYY
 
 | | Cryo-EM structure of the MukBEF monomer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ4
 
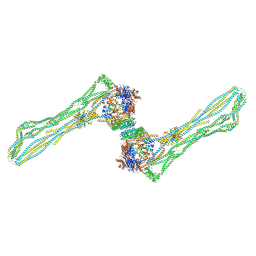 | | Cryo-EM structure of the MukBEF dimer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYX
 
 | |
7NZ2
 
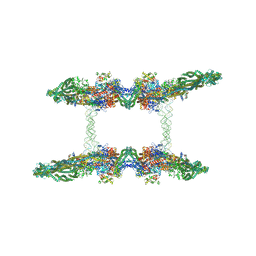 | | Cryo-EM structure of the MukBEF-MatP-DNA tetrad | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYW
 
 | | Cryo-EM structure of the MukBEF-MatP-DNA head module | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NYZ
 
 | |
7NZ0
 
 | |
7NZ3
 
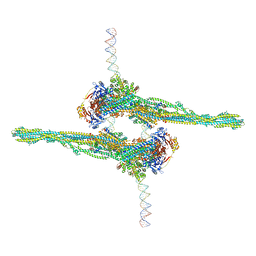 | | Cryo-EM structure of apposed MukBEF-MatP monomers on DNA | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
5N0V
 
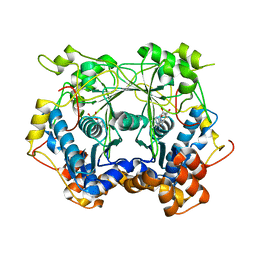 | |
5N0O
 
 | |
5N0W
 
 | |
5N0Q
 
 | |
7OGT
 
 | | Folded elbow of cohesin | | Descriptor: | Structural maintenance of chromosomes protein 1, Structural maintenance of chromosomes protein 3 | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Patele, N.J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Folding of cohesin's coiled coil is important for Scc2/4-induced association with chromosomes.
Elife, 10, 2021
|
|
5LQ0
 
 | | Crystal structure of Tyr24 phosphorylated Annexin A2 at 2.9 A resolution | | Descriptor: | Annexin A2, CALCIUM ION | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
5N0N
 
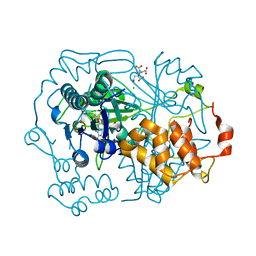 | |
5N0U
 
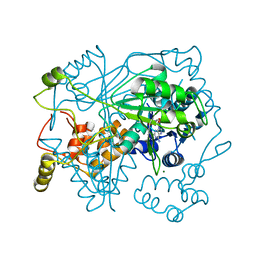 | |
5MS0
 
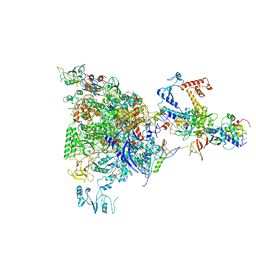 | |
5LQ2
 
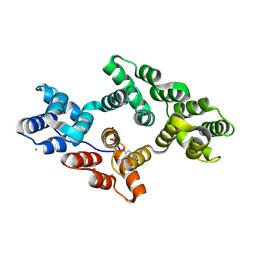 | | Crystal structure of Tyr24 phosphorylated Annexin A2 at 3.4 A resolution | | Descriptor: | Annexin A2, CALCIUM ION | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
5LM7
 
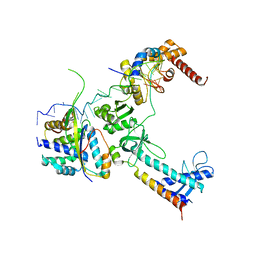 | | Crystal structure of the lambda N-Nus factor complex | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, N utilization substance protein B homolog, ... | | Authors: | Said, N, Santos, K, Weber, G, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
5N0P
 
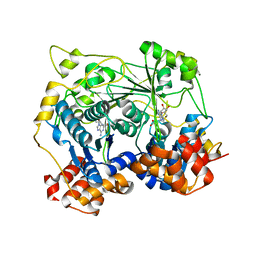 | | Crystal structure of OphA-DeltaC18 in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, peptide N-methyltranferase | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
