5FKW
 
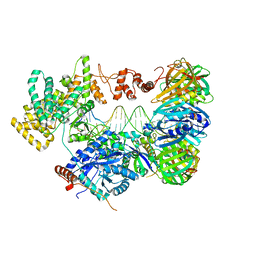 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | Descriptor: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FKV
 
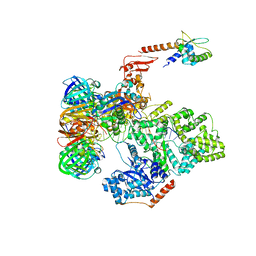 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.04 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
9FMF
 
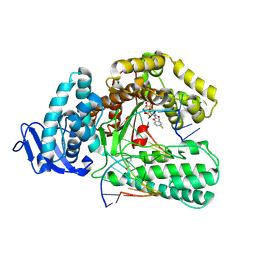 | |
7D98
 
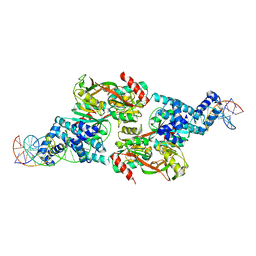 | |
4AHC
 
 | |
1DSZ
 
 | | STRUCTURE OF THE RXR/RAR DNA-BINDING DOMAIN HETERODIMER IN COMPLEX WITH THE RETINOIC ACID RESPONSE ELEMENT DR1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*G)-3'), RETINOIC ACID RECEPTOR ALPHA, ... | | Authors: | Rastinejad, F, Wagner, T, Zhao, Q, Khorasanizadeh, S. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1.
EMBO J., 19, 2000
|
|
2XZU
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND AN OXIDIZED PYRIMIDINE CONTAINING DNA AT 310K | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*VETP*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*G*A)-3', FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE, ... | | Authors: | Lebihan, Y.V, Izquierdo, M.A, Coste, F, Culard, F, Gehrke, T.H, Essalhi, K, Aller, P, Carrel, T, Castaing, B. | | Deposit date: | 2010-11-29 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 5-Hydroxy-5-Methylhydantoin DNA Lesion, a Molecular Trap for DNA Glycosylases
Nucleic Acids Res., 39, 2011
|
|
2LEV
 
 | | Structure of the DNA complex of the C-Terminal domain of Ler | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*TP*CP*AP*AP*TP*TP*AP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*AP*AP*TP*TP*GP*AP*TP*AP*GP*G)-3'), Ler | | Authors: | Cordeiro, T.N, Schimdt, H, Madrid, C, Juarez, A, Bernado, P, Grisienger, C, Garcia, J, Pons, M. | | Deposit date: | 2011-06-23 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler.
Plos Pathog., 7, 2011
|
|
4U5M
 
 | | Structure of a left-handed DNA G-quadruplex | | Descriptor: | DNA (28-MER), MAGNESIUM ION, POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Brahim, H, Chung, W.J, Lim, K.W. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a left-handed DNA G-quadruplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7PC6
 
 | | DNA-binding domain of a p53 homolog from the hydrothermal vent annelid Alvinella pompejana | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding domain, ZINC ION | | Authors: | Balourdas, D.-I, Knapp, S, Soussi, T, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evolutionary history of the p53 family DNA-binding domain: insights from an Alvinella pompejana homolog.
Cell Death Dis, 13, 2022
|
|
1TQR
 
 | | NMR Structure of DNA 17-mer GGAAAATCTCTAGCAGT corresponding to the extremity of the U5 LTR of the HIV-1 genome | | Descriptor: | DNA HIV-1 U5 LTR extremity | | Authors: | Renisio, J.G, Cosquer, S, Cherrak, I, El Antri, S, Mauffret, O, Fermandjian, S. | | Deposit date: | 2004-06-18 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Pre-organized structure of viral DNA at the binding-processing site of HIV-1 integrase
NUCLEIC ACIDS RES., 33, 2005
|
|
5XUV
 
 | | Crystal structure of DNA duplex containing 4-thiothymine-2Ag(I)-4-thiothymine base pairs | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*(8RO)P*(8RO)P*TP*CP*GP*CP*G)-3'), POTASSIUM ION, SILVER ION | | Authors: | Kondo, J, Sugawara, T, Saneyoshi, H, Ono, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a DNA duplex containing four Ag(i) ions in consecutive dinuclear Ag(i)-mediated base pairs: 4-thiothymine-2Ag(i)-4-thiothymine
Chem. Commun. (Camb.), 53, 2017
|
|
6RKW
 
 | | CryoEM structure of the complete E. coli DNA Gyrase complex bound to a 130 bp DNA duplex | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (58-MER), DNA (62-MER), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6RKS
 
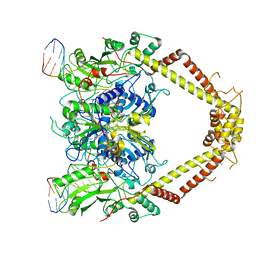 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 without TOPRIM insertion | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6RKU
 
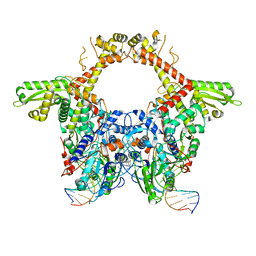 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6XEO
 
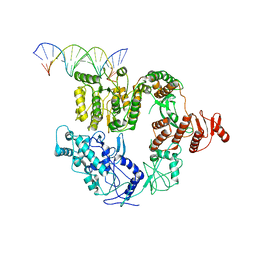 | | Structure of Mfd bound to dsDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*AP*TP*AP*CP*TP*TP*AP*CP*AP*GP*CP*CP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*GP*GP*CP*TP*GP*TP*AP*AP*GP*TP*AP*TP*CP*CP*T)-3'), Transcription-repair-coupling factor | | Authors: | Brugger, C, Deaconescu, A. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd.
Nat Commun, 11, 2020
|
|
2XZF
 
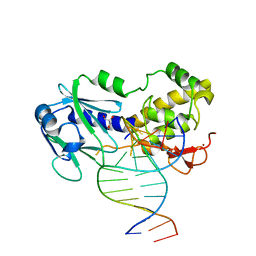 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND AN OXIDIZED PYRIMIDINE CONTAINING DNA AT 293K | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*VETP*TP*TP*TP*CP*TP*CP*GP)-3', 5'-D(*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*AP*GP*CP)-3', FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE, ... | | Authors: | LeBihan, Y.V, Izquierdo, M.A, Coste, F, Culard, F, Gehrke, T.H, Essalhi, K, Aller, P, Carrel, T, Castaing, B. | | Deposit date: | 2010-11-25 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | 5-Hydroxy-5-Methylhydantoin DNA Lesion, a Molecular Trap for DNA Glycosylases
Nucleic Acids Res., 39, 2011
|
|
6VF2
 
 | | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mg2+ (960 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mg2+ (960 min)
To be published
|
|
6VF1
 
 | | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mg2+ (120 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mg2+ (120 min)
To be published
|
|
6VF3
 
 | | DNA Polymerase Mu, 8-oxorGTP:At Ground State Ternary Complex, 50 mM Mn2+ (15 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Ground State Ternary Complex, 50 mM Mn2+ (15 min)
To be published
|
|
6VF0
 
 | | DNA Polymerase Mu, 8-oxorGTP:At Reaction State Ternary Complex, 50 mM Mg2+ (30 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Reaction State Ternary Complex, 50 mM Mg2+ (30 min)
To be published
|
|
6VF8
 
 | | DNA Polymerase Mu, 8-oxorGTP:Ct Pre-Catalytic Ternary Complex, 20 mM Ca2+ (120 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:Ct Pre-Catalytic Ternary Complex, 20 mM Ca2+ (120 min)
To be published
|
|
6VF4
 
 | | DNA Polymerase Mu, 8-oxorGTP:At Reaction State Ternary Complex, 50 mM Mn2+ (30 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Reaction State Ternary Complex, 50 mM Mn2+ (30 min)
To be published
|
|
6VEZ
 
 | | DNA Polymerase Mu, 8-oxorGTP:At Pre-Catalytic Ternary Complex, 20 mM Ca2+ (60 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.875 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Pre-Catalytic Ternary Complex, 20 mM Ca2+ (60 min)
To be published
|
|
6VF6
 
 | | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mn2+ (960 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mn2+ (960 min)
To be published
|
|
