1C21
 
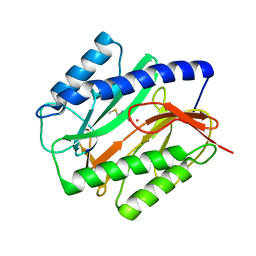 | | E. COLI METHIONINE AMINOPEPTIDASE: METHIONINE COMPLEX | | Descriptor: | COBALT (II) ION, METHIONINE, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
1C27
 
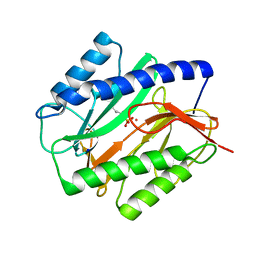 | | E. COLI METHIONINE AMINOPEPTIDASE:NORLEUCINE PHOSPHONATE COMPLEX | | Descriptor: | (1-AMINO-PENTYL)-PHOSPHONIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
2X63
 
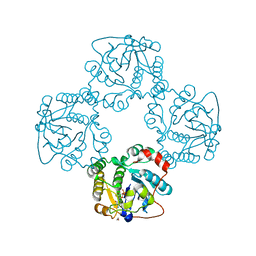 | | Crystal structure of the sialyltransferase CST-II N51A in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
2X62
 
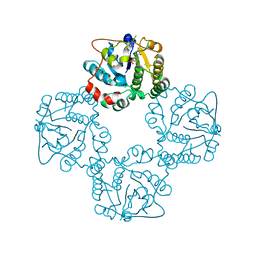 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE CST-II Y81F IN COMPLEX WITH CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, N3-PROTONATED CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
1PD5
 
 | |
6D91
 
 | | Crystal structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter in the outward-open, apo conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Divalent metal cation transporter MntH | | Authors: | Bozzi, A.T, Zimanyi, C.M, Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2018-04-27 | | Release date: | 2019-02-13 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Structures in multiple conformations reveal distinct transition metal and proton pathways in an Nramp transporter.
Elife, 8, 2019
|
|
7AEP
 
 | | Solution structure of U1-A RRM2 (190-282) | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Campagne, S, Allain, F.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | An in vitro reconstituted U1 snRNP allows the study of the disordered regions of the particle and the interactions with proteins and ligands.
Nucleic Acids Res., 49, 2021
|
|
3I3L
 
 | | Crystal structure of CmlS, a flavin-dependent halogenase | | Descriptor: | Alkylhalidase CmlS, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Podzelinska, K, Soares, A, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-06-30 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chloramphenicol Biosynthesis: The Structure of CmlS, a Flavin-Dependent Halogenase Showing a Covalent Flavin-Aspartate Bond
J.Mol.Biol., 397, 2010
|
|
1L5I
 
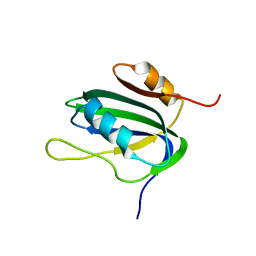 | | 30-CONFORMER NMR ENSEMBLE OF THE N-TERMINAL, DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN FROM A GEMINIVIRUS (TOMATO YELLOW LEAF CURL VIRUS-SARDINIA) | | Descriptor: | Rep protein | | Authors: | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | Deposit date: | 2002-03-07 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L2M
 
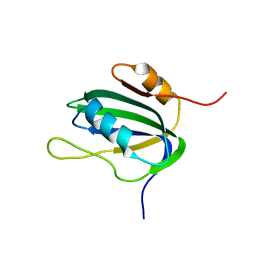 | | Minimized Average Structure of the N-terminal, DNA-binding domain of the replication initiation protein from a geminivirus (Tomato yellow leaf curl virus-Sardinia) | | Descriptor: | Rep protein | | Authors: | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | Deposit date: | 2002-02-22 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7M1K
 
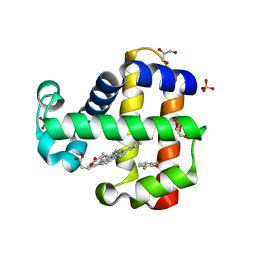 | | Crystal structure of dehaloperoxidase B in complex with 2,6-difluorophenol | | Descriptor: | 2,6-DIFLUOROPHENOL, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2021-03-13 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Bridging the functional gap between reactivity and inhibition in dehaloperoxidase B from Amphitrite ornata: Mechanistic and structural studies with 2,4- and 2,6-dihalophenols.
J.Inorg.Biochem., 236, 2022
|
|
7M1J
 
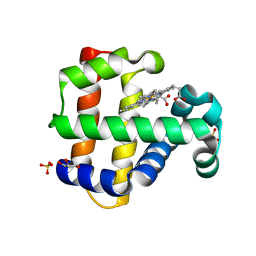 | | Crystal structure of dehaloperoxidase B in complex with 2,6-dibromophenol | | Descriptor: | 2,6-bis(bromanyl)phenol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2021-03-13 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | Bridging the functional gap between reactivity and inhibition in dehaloperoxidase B from Amphitrite ornata: Mechanistic and structural studies with 2,4- and 2,6-dihalophenols.
J.Inorg.Biochem., 236, 2022
|
|
7M1I
 
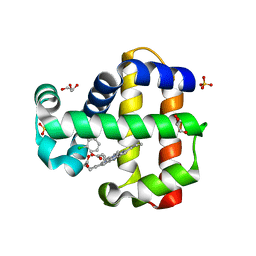 | | Crystal structure of dehaloperoxidase B in complex with 2,6-dichlorophenol | | Descriptor: | 2,6-dichlorophenol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2021-03-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Bridging the functional gap between reactivity and inhibition in dehaloperoxidase B from Amphitrite ornata: Mechanistic and structural studies with 2,4- and 2,6-dihalophenols.
J.Inorg.Biochem., 236, 2022
|
|
6U01
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84D mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, L, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5004 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
1ADX
 
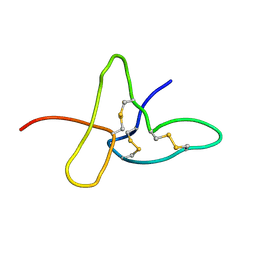 | | FIFTH EGF-LIKE DOMAIN OF THROMBOMODULIN (TMEGF5), NMR, 14 STRUCTURES | | Descriptor: | THROMBOMODULIN | | Authors: | Sampoli-Benitez, B.A, Hunter, M.J, Meininger, D.P, Komives, E.A. | | Deposit date: | 1997-02-18 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the fifth EGF-like domain of thrombomodulin: An EGF-like domain with a novel disulfide-bonding pattern.
J.Mol.Biol., 273, 1997
|
|
4NZP
 
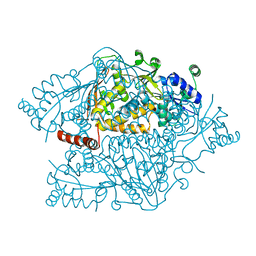 | | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Argininosuccinate synthase | | Authors: | Tan, K, Gu, M, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
4KMU
 
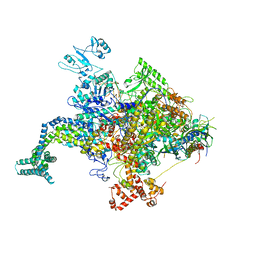 | |
1RCN
 
 | | CRYSTAL STRUCTURE OF THE RIBONUCLEASE A D(APTPAPAPG) COMPLEX : DIRECT EVIDENCE FOR EXTENDED SUBSTRATE RECOGNITION | | Descriptor: | DNA (5'-D(*AP*TP*AP*A)-3'), PROTEIN (RIBONUCLEASE A (E.C.3.1.27.5)) | | Authors: | Fontecilla-Camps, J.C, De Llorens, R, Le Du, M.H, Cuchillo, C.M. | | Deposit date: | 1994-05-27 | | Release date: | 1994-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of ribonuclease A.d(ApTpApApG) complex. Direct evidence for extended substrate recognition.
J.Biol.Chem., 269, 1994
|
|
4LY0
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTDP-Glc and 10-N-Formyl-THF | | Descriptor: | 1,2-ETHANEDIOL, 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4MLR
 
 | |
4M19
 
 | |
4LXY
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTDP and 10-N-Formyl-THF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, N-{4-[{[(6S)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}(formyl)amino]benzoyl}-L-glutamic acid, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4AJ7
 
 | | Crystallographic structure of thioredoxin from Litopenaeus vannamei (oxidized form). | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Campos-Acevedo, A.A, Sotelo-Mundo, R.R, Rudino-Pinera, E. | | Deposit date: | 2012-02-15 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | Expression, Purification, Crystallization and X-Ray Crystallographic Studies of Different Redox States of the Active Site of Thioredoxin 1 from the Whiteleg Shrimp Litopenaeus Vannamei
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3G2E
 
 | | Structure of putative OORC subunit of 2-oxoglutarate:acceptor oxidoreductase from Campylobacter jejuni | | Descriptor: | GLYCEROL, OORC subunit of 2-oxoglutarate:acceptor oxidoreductase | | Authors: | Ramagopal, U.A, Toro, R, Miller, S, Gilmore, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-31 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of putative OORC subunit of 2-oxoglutarate:acceptor oxidoreductase from
Campylobacter jejuni
To be Published
|
|
