3LWD
 
 | |
3LX8
 
 | | Crystal structure of GDP-bound NFeoB from S. thermophilus | | Descriptor: | Ferrous iron uptake transporter protein B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ash, M.R, Guilfoyle, A, Maher, M.J, Clarke, R.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B.
J.Biol.Chem., 285, 2010
|
|
3LX9
 
 | | Interconversion of Human Lysosomal Enzyme Specificities | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Tomasic, I.B, Metcalf, M.C, Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Interconversion of the specificities of human lysosomal enzymes associated with Fabry and Schindler diseases.
J.Biol.Chem., 285, 2010
|
|
3LXZ
 
 | |
3LYG
 
 | |
3LZA
 
 | |
3LR4
 
 | |
3LS5
 
 | | Anti-tetrahydrocannabinol Fab Fragment, Free Form | | Descriptor: | Heavy chain of antibody Fab fragment, Light chain of antibody Fab fragment | | Authors: | Niemi, M.H, Rouvinen, J. | | Deposit date: | 2010-02-12 | | Release date: | 2010-06-02 | | Last modified: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural insight into the molecular recognition of a (-)-Delta9-tetrahydrocannabinol and the development of a sensitive, one-step, homogeneous immunocomplex-based assay for its detection
J.Mol.Biol., 400, 2010
|
|
3LTQ
 
 | |
3LU1
 
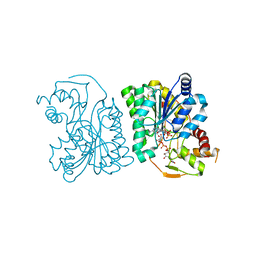 | | Crystal Structure Analysis of WbgU: a UDP-GalNAc 4-epimerase | | Descriptor: | GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bhatt, V.S, Guo, C.Y, Zhao, G, Yi, W, Liu, Z.J, Wang, P.G. | | Deposit date: | 2010-02-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Altered architecture of substrate binding region defines the unique specificity of UDP-GalNAc 4-epimerases.
Protein Sci., 20, 2011
|
|
3LUR
 
 | |
3LWF
 
 | |
3LWX
 
 | |
3LX2
 
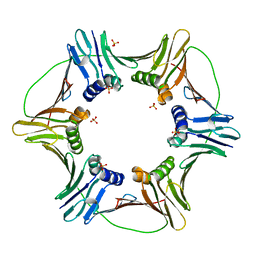 | |
3LXJ
 
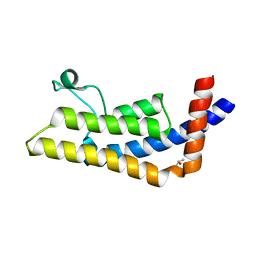 | | Crystal Structure of the Bromodomain of Human AAA domain containing 2B (ATAD2B) | | Descriptor: | ATPase family AAA domain-containing protein 2B, ISOPROPYL ALCOHOL | | Authors: | Filippakopoulos, P, Keates, T, Picaud, S, Fedorov, O, Krojer, T, Vollmar, M, Muniz, J, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3LUF
 
 | | Structure of probable two-component system response regulator/GGDEF domain protein | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, Two-component system response regulator/GGDEF domain protein | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-02 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of probable two-component system response regulator/GGDEF domain protein
To be published
|
|
3LZ0
 
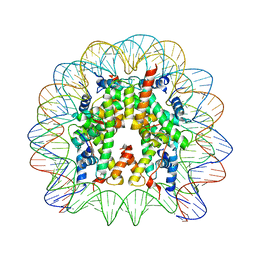 | | Crystal Structure of Nucleosome Core Particle Composed of the Widom 601 DNA Sequence (orientation 1) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A, ... | | Authors: | Vasudevan, D, Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of nucleosome core particles containing the '601' strong positioning sequence
J.Mol.Biol., 403, 2010
|
|
3LVG
 
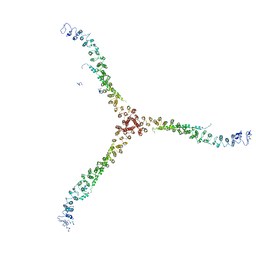 | | Crystal structure of a clathrin heavy chain and clathrin light chain complex | | Descriptor: | Clathrin heavy chain 1, Clathrin light chain B | | Authors: | Wilbur, J.D, Hwang, P.K, Ybe, J.A, Lane, M, Sellers, B.D, Jacobson, M.P, Fletterick, R.J, Brodsky, F.M. | | Deposit date: | 2010-02-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (7.94 Å) | | Cite: | Conformation switching of clathrin light chain regulates clathrin lattice assembly.
Dev.Cell, 18, 2010
|
|
3LST
 
 | | Crystal Structure of CalO1, Methyltransferase in Calicheamicin Biosynthesis, SAH bound form | | Descriptor: | 1,2-ETHANEDIOL, CalO1 Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of CalO1: a putative orsellinic acid methyltransferase in the calicheamicin-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3LX6
 
 | |
3LYK
 
 | |
3LWU
 
 | |
3LX0
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS D21N at cryogenic temperature | | Descriptor: | PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Doctrow, B.M, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-02-24 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cooperative Proton Binding in a Cluster of Carboxylic Residues
To be Published
|
|
3LZ1
 
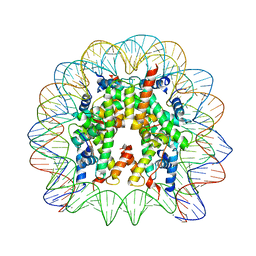 | | Crystal Structure of Nucleosome Core Particle Composed of the Widom 601 DNA Sequence (orientation 2) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A, ... | | Authors: | Vasudevan, D, Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of nucleosome core particles containing the '601' strong positioning sequence
J.Mol.Biol., 403, 2010
|
|
3LX4
 
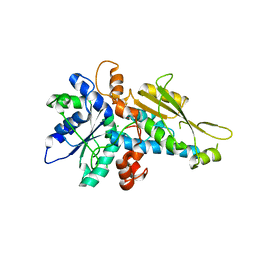 | | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(deltaEFG) | | Descriptor: | ACETATE ION, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Mulder, D.W, Boyd, E.S, Sarma, R, Lange, R.K, Endrizzi, J.A, Broderick, J.B, Peters, J.W. | | Deposit date: | 2010-02-24 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(DeltaEFG).
Nature, 465, 2010
|
|
