1J6R
 
 | |
1SEI
 
 | | STRUCTURE OF 30S RIBOSOMAL PROTEIN S8 | | Descriptor: | RIBOSOMAL PROTEIN S8 | | Authors: | Davies, C, Ramakrishnan, V, White, S.W. | | Deposit date: | 1996-08-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence for specific S8-RNA and S8-protein interactions within the 30S ribosomal subunit: ribosomal protein S8 from Bacillus stearothermophilus at 1.9 A resolution.
Structure, 4, 1996
|
|
1IFB
 
 | |
1IQ6
 
 | | (R)-HYDRATASE FROM A. CAVIAE INVOLVED IN PHA BIOSYNTHESIS | | Descriptor: | (R)-SPECIFIC ENOYL-COA HYDRATASE, ISOPROPYL ALCOHOL | | Authors: | Hisano, T, Tsuge, T, Fukui, T, Iwata, T, Doi, Y. | | Deposit date: | 2001-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the (R)-specific enoyl-CoA hydratase from Aeromonas caviae involved in polyhydroxyalkanoate
biosynthesis
J.Biol.Chem., 278, 2003
|
|
1RY5
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT FROM RHODOBACTER SPHAEROIDES WITH ASP L213 REPLACED WITH ASN | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Xu, Q, Axelrod, H.L, Abresch, E.C, Paddock, M.L, Okamura, M.Y, Feher, G. | | Deposit date: | 2003-12-19 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structure Determination of Three Mutants of the Bacterial Photosynthetic Reaction Centers from Rb. sphaeroides; Altered Proton Transfer Pathways.
STRUCTURE, 12, 2004
|
|
1IHX
 
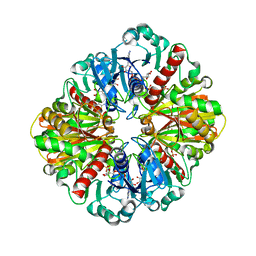 | | Crystal structure of two D-glyceraldehyde-3-phosphate dehydrogenase complexes: a case of asymmetry | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, SULFATE ION, THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shen, Y.-Q, Song, S.-Y, Lin, Z.-J. | | Deposit date: | 2001-04-20 | | Release date: | 2002-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of D-glyceraldehyde-3-phosphate dehydrogenase complexed with coenzyme analogues.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1S3M
 
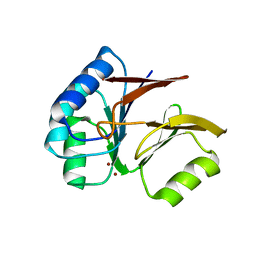 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, NICKEL (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
1ISZ
 
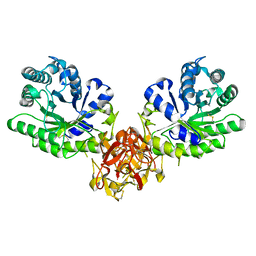 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose | | Descriptor: | beta-D-galactopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1IO2
 
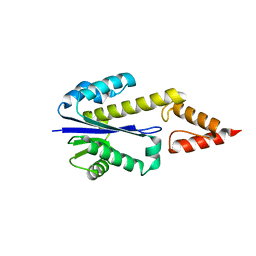 | | Crystal structure of type 2 ribonuclease h from hyperthermophilic archaeon, thermococcus kodakaraensis kod1 | | Descriptor: | RIBONUCLEASE HII | | Authors: | Muroya, A, Tsuchiya, D, Ishikawa, M, Haruki, M, Morikawa, M. | | Deposit date: | 2000-12-28 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic center of an archaeal type 2 ribonuclease H as revealed by X-ray crystallographic and mutational analyses.
Protein Sci., 10, 2001
|
|
1IUV
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-4-HYDROXYBENZOATE AT PH 5.0 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1IWO
 
 | |
1S38
 
 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2-AMINO-8-METHYLQUINAZOLIN-4(3H)-ONE | | Descriptor: | 2-AMINO-8-METHYLQUINAZOLIN-4(3H)-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Meyer, E.A, Furler, M, Diederich, F, Brenk, R, Klebe, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Synthesis and In vitro Evaluation of 2-Aminoquinazolin-4(3H)-one-based Inhibitors for tRNA-Guanine Transglycosylase (TGT)
HELV.CHIM.ACTA, 87, 2004
|
|
2BU4
 
 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
1IUD
 
 | | MALTODEXTRIN-BINDING PROTEIN INSERTION/DELETION MUTANT WITH AN INSERTED B-CELL EPITOPE FROM THE PRES2 REGION OF HEPATITIS B VIRUS | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN MALE-B133, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Saul, F.A, Vulliez-Le Normand, B, Lema, F, Bentley, G.A. | | Deposit date: | 1996-05-29 | | Release date: | 1997-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a recombinant form of the maltodextrin-binding protein carrying an inserted sequence of a B-cell epitope from the preS2 region of hepatitis B virus.
Proteins, 27, 1997
|
|
1S5P
 
 | | Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Eschericia coli. | | Descriptor: | HISTONE H4 (RESIDUES 12-19), NAD-dependent deacetylase, ZINC ION | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2004-01-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure and Substrate Binding Properties of cobB, a Sir2 Homolog Protein Deacetylase from Eschericia coli.
J.Mol.Biol., 337, 2004
|
|
2BVS
 
 | | Human thrombin complexed with fragment-based small molecules occupying the S1 pocket | | Descriptor: | ALPHA THROMBIN, HIRUDIN VARIANT-2, N-[2-(2-CARBAMOYLMETHOXY-ETHOXY)-ETHYL]-2-[2-(4-CHLORO-PHENYLSULFANYL)-ACETYLAMINO]-3-(4-GUANIDINO-PHENYL)-PROPIONAMIDE | | Authors: | Neumann, T, Junker, H.-D, Keil, O, Burkert, K, Ottleben, H, Gamer, J, Sekul, R, Deppe, H, Feurer, A, Tomandl, D, Metz, G. | | Deposit date: | 2005-07-04 | | Release date: | 2006-10-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of Thrombin Inhibitor Fragments from Chemical Microarray Screening
Lett.Drug Des.Discovery, 2, 2005
|
|
1SER
 
 | | THE 2.9 ANGSTROMS CRYSTAL STRUCTURE OF T. THERMOPHILUS SERYL-TRNA SYNTHETASE COMPLEXED WITH TRNA SER | | Descriptor: | PROTEIN (SERYL-TRNA SYNTHETASE (E.C.6.1.1.11)), TRNASER | | Authors: | Biou, S, Cusack, V, Yaremchuk, A, Tukalo, M. | | Deposit date: | 1994-02-21 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser).
Science, 263, 1994
|
|
2BXT
 
 | | Design and Discovery of Novel, Potent Thrombin Inhibitors with a Solubilizing Cationic P1-P2-Linker | | Descriptor: | 6-CHLORO-1-(2-{[(5-CHLORO-1-BENZOTHIEN-3-YL)METHYL]AMINO}ETHYL)-3-[(2-PYRIDIN-2-YLETHYL)AMINO]-1,4-DIHYDROPYRAZIN-2-OL, ALPHA THROMBIN, HIRUDIN VARIANT-2 | | Authors: | Bulat, S, Bosio, S, Grabowski, E, Papadopoulos, M.A, Cerezo-Galvez, S, Rosenbaum, C, Matassa, V.G, Ott, I, Metz, G, Schamberger, J, Sekul, R, Feurer, A. | | Deposit date: | 2005-07-27 | | Release date: | 2006-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and Discovery of Novel, Potent Pyrazinone-Based Thrombin Inhibitors with a Solubilizing P1-P2-Linker
Lett.Drug Des.Discovery, 3, 2006
|
|
1IUN
 
 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant hexagonal | | Descriptor: | ACETATE ION, meta-Cleavage product hydrolase | | Authors: | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
1J1G
 
 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1RYR
 
 | | REPLICATION OF A CIS-SYN THYMINE DIMER AT ATOMIC RESOLUTION | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3', 5'-D(*TP*TP*TP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ling, H, Boudsocq, F, Plosky, B, Woodgate, R, Yang, W. | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Replication of a Cis-Syn Thymine Dimer at Atomic Resolution
Nature, 424, 2003
|
|
1RZ2
 
 | | 1.6A crystal structure of the protein BA4783/Q81L49 (similar to sortase B) from Bacillus anthracis. | | Descriptor: | conserved hypothetical protein BA4783 | | Authors: | Wu, R, Zhang, R, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-23 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site.
Structure, 12, 2004
|
|
1S00
 
 | | PHOTOSYNTHETIC REACTION CENTER DOUBLE MUTANT FROM RHODOBACTER SPHAEROIDES WITH ASP L213 REPLACED WITH ASN AND ARG M233 REPLACED WITH CYS IN THE CHARGE-SEPARATED D+QAQB- STATE | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Xu, Q, Axelrod, H.L, Abresch, E.C, Paddock, M.L, Okamura, M.Y, Feher, G. | | Deposit date: | 2003-12-29 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure Determination of Three Mutants of the Bacterial Photosynthetic Reaction Centers from Rb. sphaeroides; Altered Proton Transfer Pathways.
STRUCTURE, 12, 2004
|
|
1J72
 
 | |
1IZ0
 
 | |
