5VBA
 
 | |
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | CHLORIDE ION, Endolysin | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5VNR
 
 | | X-ray structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5V7D
 
 | | T4 lysozyme Y18Ymbr | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Lysozyme | | Authors: | Carlsson, A.-C.C. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Increasing Enzyme Stability and Activity through Hydrogen Bond-Enhanced Halogen Bonds.
Biochemistry, 57, 2018
|
|
4I7R
 
 | |
1SWY
 
 | |
1T6H
 
 | | Crystal Structure T4 Lysozyme incorporating an unnatural amino acid p-iodo-L-phenylalanine at position 153 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Spraggon, G, Xie, J, Wang, L, Wu, N, Brock, A, Schultz, P.G. | | Deposit date: | 2004-05-06 | | Release date: | 2004-10-26 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination.
Nat.Biotechnol., 22, 2004
|
|
4I7J
 
 | | T4 Lysozyme L99A/M102H with benzene bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BENZENE, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
4I7T
 
 | |
4I7O
 
 | |
4I7M
 
 | | T4 Lysozyme L99A/M102H with 2-allylphenol bound | | Descriptor: | 2-ALLYLPHENOL, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
4I7N
 
 | | T4 Lysozyme L99A/M102H with 1-phenyl-2-propyn-1-ol bound | | Descriptor: | (1R)-1-phenylprop-2-yn-1-ol, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
4I7P
 
 | | T4 Lysozyme L99A/M102H with 4-bromoimidazole bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-bromo-1H-imidazole, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
1SSW
 
 | | Crystal structure of phage T4 lysozyme mutant Y24A/Y25A/T26A/I27A/C54T/C97A | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
1SX2
 
 | |
1TLA
 
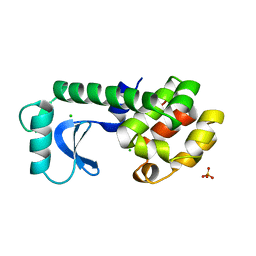 | | HYDROPHOBIC CORE REPACKING AND AROMATIC-AROMATIC INTERACTION IN THE THERMOSTABLE MUTANT OF T4 LYSOZYME SER 117 (RIGHT ARROW) PHE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, T4 LYSOZYME | | Authors: | Anderson, D.E, Hurley, J.H, Nicholson, H, Baase, W.A, Matthews, B.W. | | Deposit date: | 1993-03-22 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophobic core repacking and aromatic-aromatic interaction in the thermostable mutant of T4 lysozyme Ser 117-->Phe.
Protein Sci., 2, 1993
|
|
1SWZ
 
 | |
1T8G
 
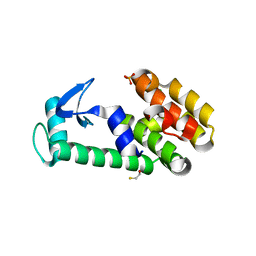 | | Crystal structure of phage T4 lysozyme mutant L32A/L33A/T34A/C54T/C97A/E108V | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | He, M.M, Wood, Z.A, Baase, W.A, Xiao, H, Matthews, B.W. | | Deposit date: | 2004-05-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation.
Protein Sci., 13, 2004
|
|
4I7K
 
 | | T4 Lysozyme L99A/M102H with toluene bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
4I7Q
 
 | |
4I7L
 
 | | T4 Lysozyme L99A/M102H with phenol bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
4I7S
 
 | |
1SX7
 
 | |
1SSY
 
 | | Crystal structure of phage T4 lysozyme mutant G28A/I29A/G30A/C54T/C97A | | Descriptor: | Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
1T8F
 
 | | Crystal structure of phage T4 lysozyme mutant R14A/K16A/I17A/K19A/T21A/E22A/C54T/C97A | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | He, M.M, Wood, Z.A, Baase, W.A, Xiao, H, Matthews, B.W. | | Deposit date: | 2004-05-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation.
Protein Sci., 13, 2004
|
|
