7ADR
 
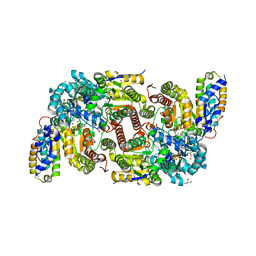 | | CO bound as bridging ligand at the active site of vanadium nitrogenase VFe protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Grunau, K, Einsle, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7CSN
 
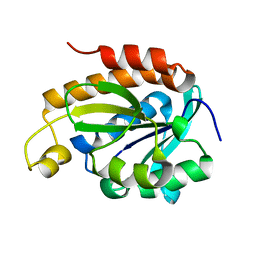 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.00 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-08-15 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.00 A resolution
To Be Published
|
|
6ODG
 
 | | SVQIVY, Crystal Structure of a tau protein fragment | | Descriptor: | Microtubule-associated protein tau | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Seidler, P.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure-based inhibitors halt prion-like seeding by Alzheimer's disease-and tauopathy-derived brain tissue samples.
J.Biol.Chem., 294, 2019
|
|
5RT2
 
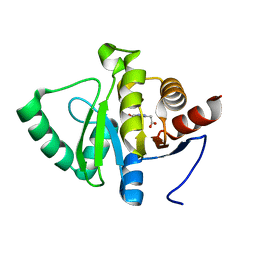 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000008652361 | | Descriptor: | 2-(4-oxidanylidene-3~{H}-phthalazin-1-yl)ethanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTW
 
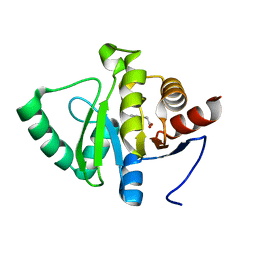 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000164777 | | Descriptor: | (2-HYDROXYPHENYL)ACETIC ACID, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUC
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000005878 | | Descriptor: | NICOTINAMIDE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUR
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000017744334 | | Descriptor: | 6-fluoro-1,3-benzothiazol-2-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RV7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000003954002 | | Descriptor: | 1H-indazol-3-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4W5L
 
 | | Crystal structure of a prp peptide | | Descriptor: | PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-18 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
2RBK
 
 | |
6RI0
 
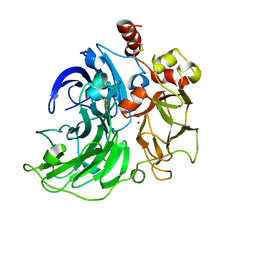 | | Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Ninth structure of the series with 1215 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1OAI
 
 | |
4X6H
 
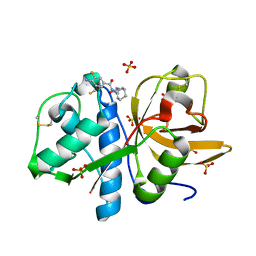 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | Descriptor: | 4-amino-3-fluoro-N-(1-{[(2Z)-2-iminoethyl]carbamoyl}cyclohexyl)benzamide, 4-amino-N-{1-[(cyanomethyl)carbamoyl]cyclohexyl}-3-fluorobenzamide, Cathepsin K, ... | | Authors: | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
5RBX
 
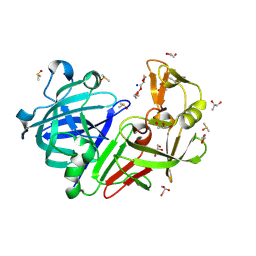 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library D10a | | Descriptor: | 2-methyl-N-(4-methylphenyl)-L-alanine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCF
 
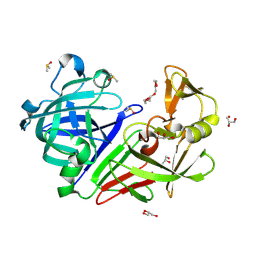 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library H10b | | Descriptor: | (1R)-1-(4-fluorophenyl)-N-[2-(1H-pyrazol-1-yl)ethyl]ethan-1-amine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1MSO
 
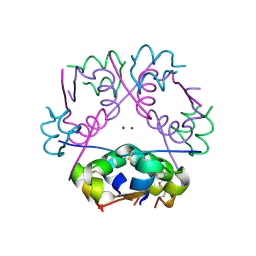 | | T6 Human Insulin at 1.0 A Resolution | | Descriptor: | Insulin A-Chain, Insulin B-Chain, ZINC ION | | Authors: | Smith, G.D, Pangborn, W.A, Blessing, R.H. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The structure of T6 human insulin at 1.0 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
8DPY
 
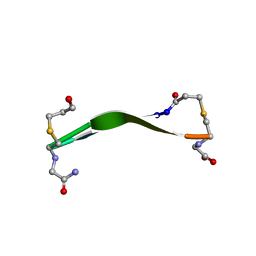 | |
5XBU
 
 | | Crystal structure of GH45 endoglucanase EG27II in apo-form | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1EB6
 
 | | Deuterolysin from Aspergillus oryzae | | Descriptor: | 1,2-ETHANEDIOL, NEUTRAL PROTEASE II, ZINC ION | | Authors: | McAuley, K.E, Jia-Xing, Y, Dodson, E.J, Lehmbeck, J, Ostergaard, P.R, Wilson, K.S. | | Deposit date: | 2001-07-19 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Quick Solution: Ab Initio Structure Determination of a 19 kDa Metalloproteinase Using Acorn
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5RSU
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000002055 | | Descriptor: | Non-structural protein 3, salicylamide | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3E6Z
 
 | |
5RTS
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000159004 | | Descriptor: | 5-phenylpyridine-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RU8
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000154817 | | Descriptor: | ISOQUINOLIN-1-AMINE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4UBY
 
 | |
5RUP
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000004976927 | | Descriptor: | Non-structural protein 3, [3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]acetic acid | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
