4FZV
 
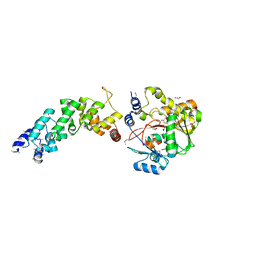 | | Crystal structure of the human MTERF4:NSUN4:SAM ternary complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative methyltransferase NSUN4, ... | | Authors: | Guja, K.E, Yakubovskaya, E, Mejia, E, Castano, S, Hambardjieva, E, Choi, W.S, Garcia-Diaz, M. | | Deposit date: | 2012-07-08 | | Release date: | 2012-10-03 | | Last modified: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.9996 Å) | | Cite: | Structure of the Essential MTERF4:NSUN4 Protein Complex Reveals How an MTERF Protein Collaborates to Facilitate rRNA Modification.
Structure, 20, 2012
|
|
4CRM
 
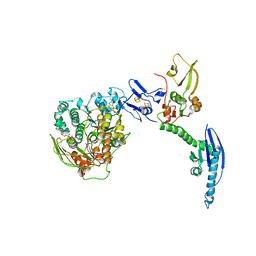 | | Cryo-EM of a pre-recycling complex with eRF1 and ABCE1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, EUKARYOTIC PEPTIDE CHAIN RELEASE FACTOR SUBUNIT 1, ... | | Authors: | Preis, A, Heuer, A, Barrio-Garcia, C, Hauser, A, Eyler, D, Berninghausen, O, Green, R, Becker, T, Beckmann, R. | | Deposit date: | 2014-02-28 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.75 Å) | | Cite: | Cryoelectron Microscopic Structures of Eukaryotic Translation Termination Complexes Containing Erf1-Erf3 or Erf1-Abce1.
Cell Rep., 8, 2014
|
|
2D10
 
 | |
3ZGQ
 
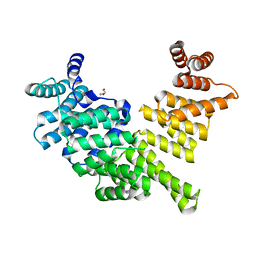 | | Crystal structure of human interferon-induced protein IFIT5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, INTERFERON-INDUCED PROTEIN WITH TETRATRICOPEPTIDE REPEATS 5 | | Authors: | Katibah, G.E, Lee, H.J, Huizar, J.P, Vogan, J.M, Alber, T, Collins, K. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-23 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | TRNA Binding, Structure, and Localization of the Human Interferon-Induced Protein Ifit5.
Mol.Cell, 49, 2013
|
|
2LLX
 
 | |
3PIW
 
 | | Zebrafish interferon 2 | | Descriptor: | Type I interferon 2 | | Authors: | Hamming, O.J, Hartmann, R, Lutfalla, G, Levraud, J.-P. | | Deposit date: | 2010-11-08 | | Release date: | 2011-07-20 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Crystal Structure of Zebrafish Interferons I and II Reveals Conservation of Type I Interferon Structure in Vertebrates.
J.Virol., 85, 2011
|
|
1RFB
 
 | |
1RH2
 
 | | RECOMBINANT HUMAN INTERFERON-ALPHA 2B | | Descriptor: | INTERFERON-ALPHA 2B, ZINC ION | | Authors: | Walter, M.R. | | Deposit date: | 1996-11-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Zinc mediated dimer of human interferon-alpha 2b revealed by X-ray crystallography.
Structure, 4, 1996
|
|
1DT9
 
 | |
2KTU
 
 | |
1IF1
 
 | | INTERFERON REGULATORY FACTOR 1 (IRF-1) COMPLEX WITH DNA | | Descriptor: | DNA (26-MER), PROTEIN (INTERFERON REGULATORY FACTOR 1) | | Authors: | Escalante, C.R, Yie, J, Thanos, D, Aggarwal, A. | | Deposit date: | 1997-09-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of IRF-1 with bound DNA reveals determinants of interferon regulation.
Nature, 391, 1998
|
|
2JXO
 
 | | Structure of the second PDZ domain of NHERF-1 | | Descriptor: | Ezrin-radixin-moesin-binding phosphoprotein 50 | | Authors: | Cheng, H, Li, J, Dai, Z, Bu, Z, Roder, H. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory Interactions between the PDZ2 and C-terminal Domains in the Scaffolding Protein NHERF1
Structure, 17, 2009
|
|
2KZ1
 
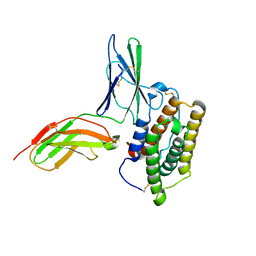 | | Inter-molecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric back-protonation and 2D NOESY | | Descriptor: | Interferon alpha-2, Soluble IFN alpha/beta receptor | | Authors: | Nudelman, I, Akabayov, S.R, Schnur, E, Biron, Z, Levy, R, Xu, Y, Yang, D, Anglister, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | Intermolecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric reverse-protonation and two-dimensional NOESY
Biochemistry, 49, 2010
|
|
2O6G
 
 | |
1CYI
 
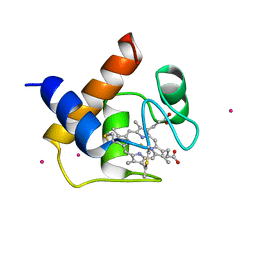 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
1CYJ
 
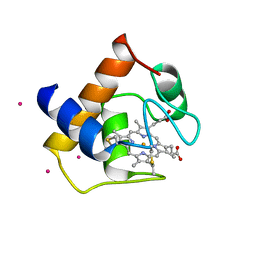 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
5ONI
 
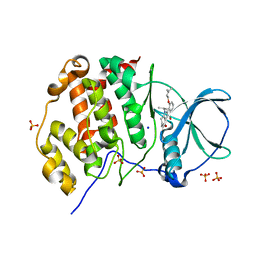 | | LOW-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,4-BUTANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
5OOI
 
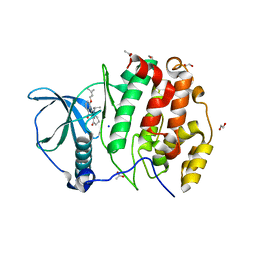 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA') IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, ACETATE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-07 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
5OMY
 
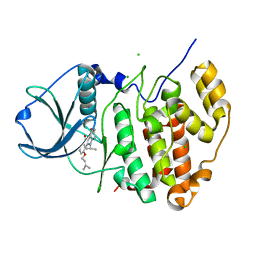 | | HIGH-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-02 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
5WIR
 
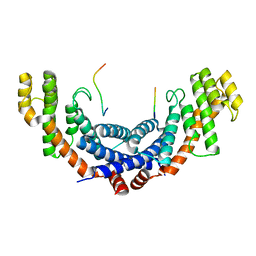 | | Structure of the TRF1-TERB1 interface | | Descriptor: | TERB1-TBM, Telomeric repeat-binding factor 1 | | Authors: | Nandakumar, J, Pendlebury, D.F, Smith, E.M, Tesmer, V.M. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dissecting the telomere-inner nuclear membrane interface formed in meiosis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1OIX
 
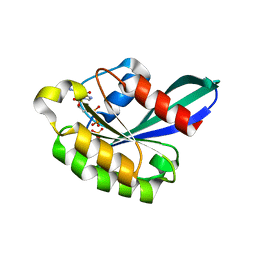 | | X-ray structure of the small G protein Rab11a in complex with GDP and Pi | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pasqualato, S, Senic-Matuglia, F, Renault, L, Goud, B, Salamero, J, Cherfils, J. | | Deposit date: | 2003-06-26 | | Release date: | 2005-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Evidence for Substrate-Assisted GTP Hydrolysis by a Small GTP Binding Protein
Structure, 13, 2005
|
|
4LQR
 
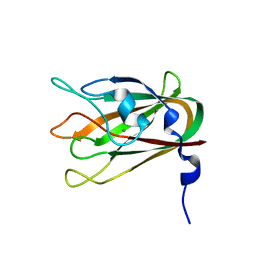 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein | | Authors: | Grondin, J.M, Furness, H.S, Duan, D, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
3L31
 
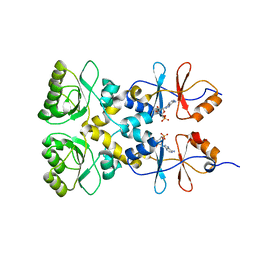 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with the inhibitor, AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
1M13
 
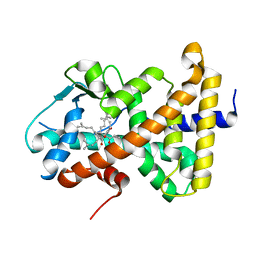 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
8UB5
 
 | | The Apo NanH structure from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, Sialidase | | Authors: | Medley, B.J, Boraston, A.B. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A "terminal" case of glycan catabolism: structural and enzymatic characterization of the sialidases of Clostridium perfringens.
J.Biol.Chem., 2024
|
|
