5W2K
 
 | |
5W2R
 
 | |
5W2X
 
 | |
5W31
 
 | |
5W3B
 
 | |
3QDL
 
 | | Crystal structure of RdxA from Helicobacter pyroli | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Oxygen-insensitive NADPH nitroreductase | | Authors: | Rojas, A.L, Martinez-Julvez, M, Olekhnovich, I.N, Hoffman, P.S, Sancho, J. | | Deposit date: | 2011-01-18 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of RdxA--an oxygen-insensitive nitroreductase essential for metronidazole activation in Helicobacter pylori.
Febs J., 279, 2012
|
|
5W37
 
 | |
3QSI
 
 | |
3PHH
 
 | | Shikimate 5-Dehydrogenase (aroE) from Helicobacter pylori in complex with Shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | Authors: | Cheng, W.C, Lin, S.C, Wang, W.C. | | Deposit date: | 2010-11-04 | | Release date: | 2011-11-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Shikimate 5-Dehydrogenase (aroE) from Helicobacter pylori in complex with Shikimate
To be Published
|
|
3PHG
 
 | |
3PHI
 
 | | Shikimate 5-Dehydrogenase (aroE) from Helicobacter pylori in complex with Shikimate and NADPH | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Shikimate dehydrogenase | | Authors: | Cheng, W.C, Lin, S.C, Wang, W.C. | | Deposit date: | 2010-11-04 | | Release date: | 2011-11-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Shikimate 5-Dehydrogenase (aroE) from Helicobacter pylori in complex with Shikimate and NADPH
To be Published
|
|
3QXH
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ADP and 8-aminocaprylic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Minor, C, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXJ
 
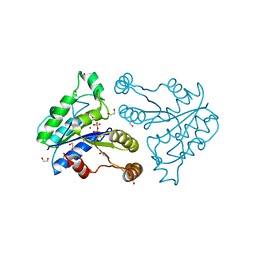 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GTP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QY0
 
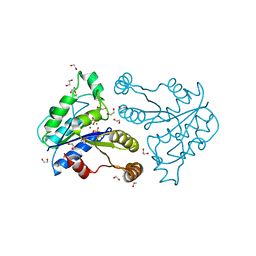 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QGA
 
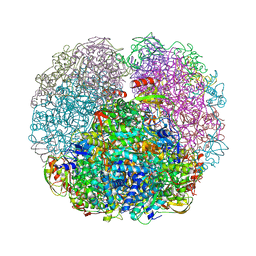 | |
3OQG
 
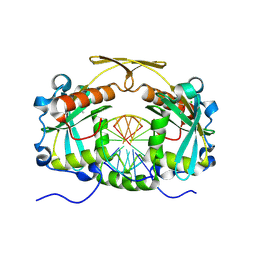 | | Restriction endonuclease HPY188I in complex with substrate DNA | | Descriptor: | CHLORIDE ION, DNA 5'-D(*GP*AP*TP*CP*TP*GP*AP*AP*C)-3', DNA 5'-D(*GP*TP*TP*CP*AP*GP*AP*TP*C)-3', ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2010-09-03 | | Release date: | 2010-10-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hpy188I-DNA pre- and post-cleavage complexes--snapshots of the GIY-YIG nuclease mediated catalysis.
Nucleic Acids Res., 39, 2011
|
|
5W39
 
 | | Crystal structure of mutant CJ YCEI protein (CJ-N182C) with monobromobimane guest structure | | Descriptor: | 3-(bromomethyl)-2,5,6-trimethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, EICOSANE, Polyisoprenoid-binding protein, ... | | Authors: | Huber, T.R, Snow, C.D. | | Deposit date: | 2017-06-07 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Installing Guest Molecules at Specific Sites within Scaffold Protein Crystals.
Bioconjug. Chem., 29, 2018
|
|
5W3A
 
 | |
3QGK
 
 | |
3QXX
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GDP and 8-aminocaprylic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-aminooctanoic acid, Dethiobiotin synthetase, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
1R5G
 
 | | Crystal Structure of MetAP2 complexed with A311263 | | Descriptor: | (2S,3R)-3-AMINO-2-HYDROXY-5-(ETHYLSULFANYL)PENTANOYL-((S)-(-)-(1-NAPHTHYL)ETHYL)AMIDE, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
3O1Q
 
 | |
1QXW
 
 | | Crystal structure of Staphyloccocus aureus in complex with an aminoketone inhibitor 54135. | | Descriptor: | (3S)-3-AMINO-1-(CYCLOPROPYLAMINO)HEPTANE-2,2-DIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
3PHT
 
 | |
7KK1
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site and L-lysine bound at the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-27 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A TIGHT DIMER INTERFACE N84 RESIDUE, PLAYS A CRITICAL ROLE IN THE TRANSMISSION OF THE ALLOSTERIC INHIBITION SIGNALS IN Cj.DHDPS
To Be Published
|
|
