5W2Z
 
 | |
1LAM
 
 | | LEUCINE AMINOPEPTIDASE (UNLIGATED) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CARBONATE ION, LEUCINE AMINOPEPTIDASE, ... | | Authors: | Straeter, N, Lipscomb, W.N. | | Deposit date: | 1995-08-11 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two-metal ion mechanism of bovine lens leucine aminopeptidase: active site solvent structure and binding mode of L-leucinal, a gem-diolate transition state analogue, by X-ray crystallography.
Biochemistry, 34, 1995
|
|
1LCP
 
 | |
3FMR
 
 | | Crystal structure of an Encephalitozoon cuniculi methionine aminopeptidase type 2 with angiogenesis inhibitor TNP470 bound | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, FE (III) ION, Methionine aminopeptidase 2, ... | | Authors: | Alvarado, J.J, Russell, M, Zhang, A, Adams, J, Toro, R, Burley, S.K, Weiss, L.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470.
Mol.Biochem.Parasitol., 168, 2009
|
|
5W37
 
 | |
5W39
 
 | | Crystal structure of mutant CJ YCEI protein (CJ-N182C) with monobromobimane guest structure | | Descriptor: | 3-(bromomethyl)-2,5,6-trimethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, EICOSANE, Polyisoprenoid-binding protein, ... | | Authors: | Huber, T.R, Snow, C.D. | | Deposit date: | 2017-06-07 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Installing Guest Molecules at Specific Sites within Scaffold Protein Crystals.
Bioconjug. Chem., 29, 2018
|
|
5W3A
 
 | |
2V3Z
 
 | |
1LAP
 
 | | MOLECULAR STRUCTURE OF LEUCINE AMINOPEPTIDASE AT 2.7-ANGSTROMS RESOLUTION | | Descriptor: | Cytosol aminopeptidase, ZINC ION | | Authors: | Burley, S.K, David, P.R, Taylor, A, Lipscomb, W.N. | | Deposit date: | 1990-08-01 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular structure of leucine aminopeptidase at 2.7-A resolution.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
4NET
 
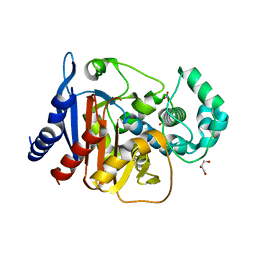 | | Crystal structure of ADC-1 beta-lactamase | | Descriptor: | AmpC, GLYCEROL, NITRATE ION | | Authors: | Bhattacharya, M, Toth, M, Antunes, N.T, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the extended-spectrum class C beta-lactamase ADC-1 from Acinetobacter baumannii.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3H8G
 
 | | Bestatin complex structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, BICARBONATE ION, Cytosol aminopeptidase, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
3H8E
 
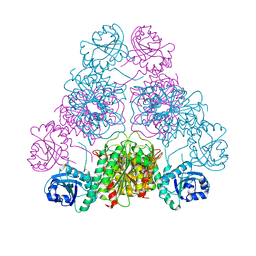 | |
5LG6
 
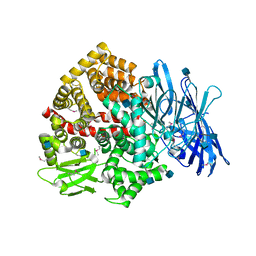 | | Structure of the deglycosylated porcine aminopeptidase N ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ZINC ION | | Authors: | Santiago, C, Reguera, J, Mudgal, G, Casasnovas, J.M. | | Deposit date: | 2016-07-06 | | Release date: | 2017-04-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric inhibition of aminopeptidase N functions related to tumor growth and virus infection.
Sci Rep, 7, 2017
|
|
2V3X
 
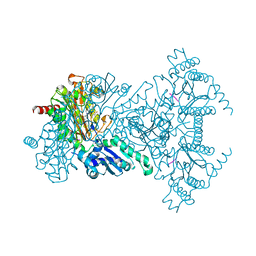 | |
2V3Y
 
 | |
6GMM
 
 | | Crystal structure of Helicobacter pylori adhesin LabA | | Descriptor: | adhesin LabA | | Authors: | Paraskevopoulou, V, Overman, R.C, Stolnik, S, Winkler, S, Gellert, P, Falcone, F.H, Schimpl, M. | | Deposit date: | 2018-05-27 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and binding characterization of the LacdiNAc-specific adhesin (LabA; HopD) exodomain from Helicobacter pylori
Current Research in Structural Biology, 2021
|
|
5W2D
 
 | |
5W30
 
 | | Crystal structure of mutant CJ YCEI protein (CJ-N48C) with monobromobimane guest structure | | Descriptor: | 3-(bromomethyl)-2,5,6-trimethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, Putative periplasmic protein, SULFATE ION, ... | | Authors: | Huber, T.R, Snow, C.D. | | Deposit date: | 2017-06-07 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Installing Guest Molecules at Specific Sites within Scaffold Protein Crystals.
Bioconjug. Chem., 29, 2018
|
|
6GBH
 
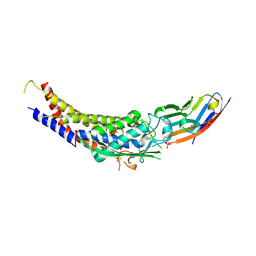 | | Helicobacter pylori adhesin HopQ type II bound to the N-terminal domain of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, HopQ, SULFATE ION | | Authors: | Moonens, K, Kruse, T, Gerhard, M, Remaut, H. | | Deposit date: | 2018-04-13 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Helicobacter pyloriadhesin HopQ disruptstransdimerization in human CEACAMs.
EMBO J., 37, 2018
|
|
5LHD
 
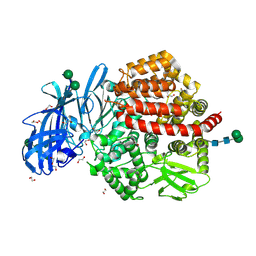 | | Structure of glycosylated human aminopeptidase N | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Mudgal, G, Santiago, C, Casasnovas, J.M. | | Deposit date: | 2016-07-11 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric inhibition of aminopeptidase N functions related to tumor growth and virus infection.
Sci Rep, 7, 2017
|
|
5LDS
 
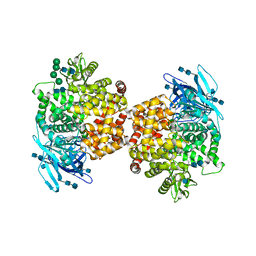 | | Structure of the porcine aminopeptidase N ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Santiago, C, Reguera, J, Mudgal, G, Casasnovas, J.M. | | Deposit date: | 2016-06-27 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of aminopeptidase N functions related to tumor growth and virus infection.
Sci Rep, 7, 2017
|
|
5LZN
 
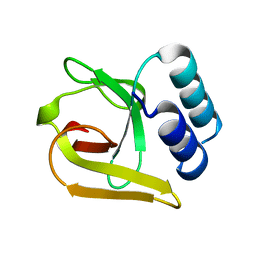 | | -TIP microtubule-binding domain | | Descriptor: | Calmodulin-regulated spectrin-associated protein 3 | | Authors: | Stangier, M.M, Steinmetz, M.O. | | Deposit date: | 2016-09-30 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2Q95
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor A05 | | Descriptor: | 5-(2-CHLORO-4-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
3H8F
 
 | | High pH native structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | BICARBONATE ION, Cytosol aminopeptidase, MANGANESE (II) ION, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
2Q92
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor B23 | | Descriptor: | 5-(2-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
