8XYZ
 
 | | The structure of fox ACE2 and PT RBD complex | | Descriptor: | Angiotensin-converting enzyme, Signal peptide, Spike protein S1, ... | | Authors: | sun, J.Q. | | Deposit date: | 2024-01-20 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The structure of fox ACE2/PT RBD complex
To Be Published
|
|
8XYB
 
 | | hPhK gamma-delta subcomplex in inactive state | | Descriptor: | Calmodulin-1, FARNESYL, Phosphorylase b kinase gamma catalytic chain, ... | | Authors: | Yang, X.K, Xiao, J.Y. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture and activation of human muscle phosphorylase kinase.
Nat Commun, 15, 2024
|
|
8XYA
 
 | |
8XY7
 
 | | hPhK alpha-gamma subcomplex in active state | | Descriptor: | FARNESYL, Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform, ... | | Authors: | Yang, X.K, Xiao, J.Y. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture and activation of human muscle phosphorylase kinase.
Nat Commun, 15, 2024
|
|
8XY0
 
 | | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Endo-1,4-beta-xylanase A | | Authors: | Nakamura, T, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | Deposit date: | 2024-01-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
8XXW
 
 | | Fab M2-7 complexed with SARS-Cov2 RBD and human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, M2-7-Heavy chain, M2-7-Light chain, ... | | Authors: | Liu, C, Xie, Y. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Mosaic RBD nanoparticle elicits immunodominant antibody responses across sarbecoviruses.
Cell Rep, 43, 2024
|
|
8XXN
 
 | | Cryo-EM structure of the human 43S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXM
 
 | | Cryo-EM structure of the human 40S ribosome with PDCD4 and eIF3G | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXL
 
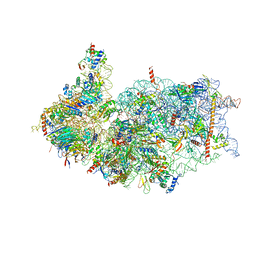 | | Cryo-EM structure of the human 40S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXB
 
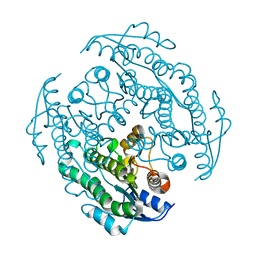 | |
8XXA
 
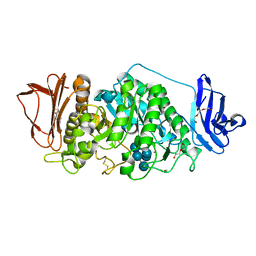 | |
8XX9
 
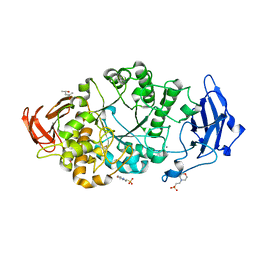 | | Rhodothermus marinus alpha-amylase RmGH13_47A CBM48-A-B-C domains | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2024-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of alpha-1,6-branched alpha-glucan by GH13_47 alpha-amylase from Rhodothermus marinus.
Proteins, 92, 2024
|
|
8XX1
 
 | |
8XWX
 
 | |
8XWK
 
 | |
8XW4
 
 | | Cryo-EM structure of TMEM63B-Digitonin state | | Descriptor: | CSC1-like protein 2 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XW3
 
 | |
8XW2
 
 | |
8XW1
 
 | | Cryo-EM structure of OSCA1.2-V335W-DDM state | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.49 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XW0
 
 | | Cryo-EM structure of OSCA3.1-GDN state | | Descriptor: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XVZ
 
 | |
8XVY
 
 | |
8XVX
 
 | |
8XVF
 
 | |
8XV9
 
 | | Fedratinib-bound human SLC19A3 | | Descriptor: | N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide, Soluble cytochrome b562,Thiamine transporter 2 | | Authors: | Dang, Y, Wang, G.P, Zhang, Z. | | Deposit date: | 2024-01-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Substrate and drug recognition mechanisms of SLC19A3.
Cell Res., 34, 2024
|
|
