1HRQ
 
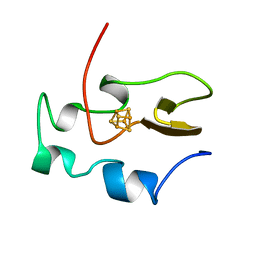 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE REDUCED HIGH-POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-01-17 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
1HRR
 
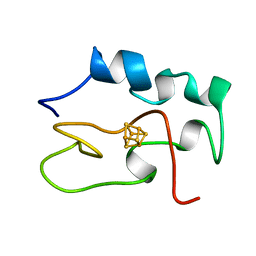 | | THE THREE DIMENSIONAL STRUCTURE OF THE REDUCED HIGH POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | Descriptor: | IRON/SULFUR CLUSTER, REDUCED HIGH POTENTIAL IRON SULFUR PROTEIN | | Authors: | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-01-17 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
1HRS
 
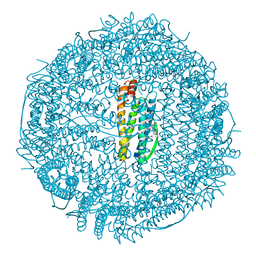 | | A CRYSTALLOGRAPHIC STUDY OF HAEM BINDING TO FERRITIN | | Descriptor: | APOFERRITIN CO-CRYSTALLIZED WITH SN-PROTOPORPHYRIN IX IN CADMIUM SULFATE, CADMIUM ION, PROTOPORPHYRIN IX | | Authors: | Precigoux, G, Yariv, J, Gallois, B, Dautant, A, Courseille, C, Langlois D'Estaintot, B. | | Deposit date: | 1993-11-05 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of haem binding to ferritin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1HRT
 
 | |
1HRU
 
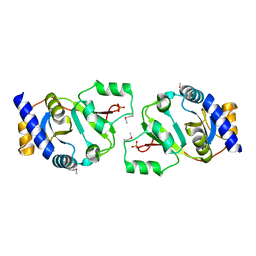 | | THE STRUCTURE OF THE YRDC GENE PRODUCT FROM E.COLI | | Descriptor: | PHOSPHATE ION, YRDC GENE PRODUCT | | Authors: | Teplova, M, Tereshko, V, Sanishvili, R, Joachimiak, A, Bushueva, T, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding.
Protein Sci., 9, 2000
|
|
1HRV
 
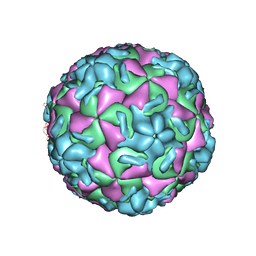 | | HRV14/SDZ 35-682 COMPLEX | | Descriptor: | 1-[2-HYDROXY-3-(4-CYCLOHEXYL-PHENOXY)-PROPYL]-4-(2-PYRIDYL)-PIPERAZINE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Oren, D.A, Zhang, A, Arnold, E. | | Deposit date: | 1995-03-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | SDZ 35-682, a new picornavirus capsid-binding agent with potent antiviral activity.
Antiviral Res., 26, 1995
|
|
1HRY
 
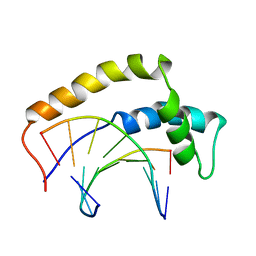 | | THE 3D STRUCTURE OF THE HUMAN SRY-DNA COMPLEX SOLVED BY MULTID-DIMENSIONAL HETERONUCLEAR-EDITED AND-FILTERED NMR | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*GP*TP*GP*C)-3'), HUMAN SRY | | Authors: | Clore, G.M, Werner, M.H, Huth, J.R, Gronenborn, A.M. | | Deposit date: | 1995-05-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex.
Cell(Cambridge,Mass.), 81, 1995
|
|
1HRZ
 
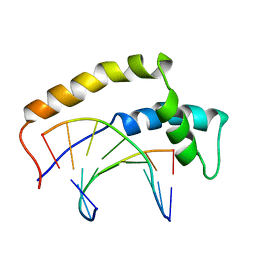 | | THE 3D STRUCTURE OF THE HUMAN SRY-DNA COMPLEX SOLVED BY MULTI-DIMENSIONAL HETERONUCLEAR-EDITED AND-FILTERED NMR | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*GP*TP*GP*C)-3'), HUMAN SRY | | Authors: | Clore, G.M, Werner, M.H, Huth, J.R, Gronenborn, A.M. | | Deposit date: | 1995-05-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex.
Cell(Cambridge,Mass.), 81, 1995
|
|
1HS1
 
 | |
1HS2
 
 | |
1HS3
 
 | |
1HS4
 
 | |
1HS5
 
 | | NMR SOLUTION STRUCTURE OF DESIGNED P53 DIMER | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Davison, T.S, Nie, X, Ma, W, Li, Y, Kay, C, Benchimol, S, Arrowsmith, C.H. | | Deposit date: | 2000-12-22 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and functionality of a designed p53 dimer.
J.Mol.Biol., 307, 2001
|
|
1HS6
 
 | | STRUCTURE OF LEUKOTRIENE A4 HYDROLASE COMPLEXED WITH BESTATIN. | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, ACETATE ION, IMIDAZOLE, ... | | Authors: | Thunnissen, M.M.G.M, Nordlund, P.N, Haeggstrom, J.Z. | | Deposit date: | 2000-12-24 | | Release date: | 2001-06-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human leukotriene A(4) hydrolase, a bifunctional enzyme in inflammation.
Nat.Struct.Biol., 8, 2001
|
|
1HS7
 
 | | VAM3P N-TERMINAL DOMAIN SOLUTION STRUCTURE | | Descriptor: | SYNTAXIN VAM3 | | Authors: | Dulubova, I, Yamaguchi, T, Wang, Y, Sudhof, T.C, Rizo, J. | | Deposit date: | 2000-12-24 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Vam3p structure reveals conserved and divergent properties of syntaxins.
Nat.Struct.Biol., 8, 2001
|
|
1HS8
 
 | |
1HSA
 
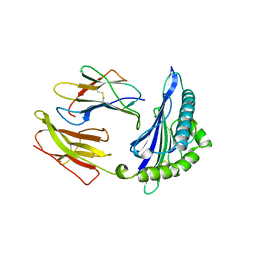 | | THE THREE-DIMENSIONAL STRUCTURE OF HLA-B27 AT 2.1 ANGSTROMS RESOLUTION SUGGESTS A GENERAL MECHANISM FOR TIGHT PEPTIDE BINDING TO MHC | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-B*2705), MODEL PEPTIDE SEQUENCE - ARAAAAAAA | | Authors: | Madden, D.R, Gorga, J.C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1992-08-11 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC.
Cell(Cambridge,Mass.), 70, 1992
|
|
1HSB
 
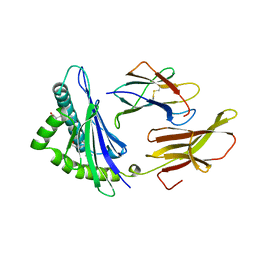 | | DIFFERENT LENGTH PEPTIDES BIND TO HLA-AW68 SIMILARLY AT THEIR ENDS BUT BULGE OUT IN THE MIDDLE | | Descriptor: | ALANINE, ARGININE, BOUND PEPTIDE FRAGMENT, ... | | Authors: | Guo, H.-C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1993-03-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle.
Nature, 360, 1992
|
|
1HSE
 
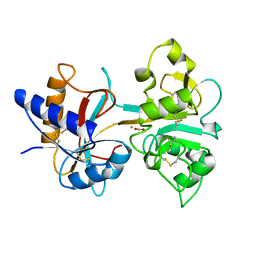 | | H253M N TERMINAL LOBE OF HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Nicholson, H, Anderson, B.F, Baker, E.N. | | Deposit date: | 1996-12-11 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis of the histidine ligand in human lactoferrin: iron binding properties and crystal structure of the histidine-253-->methionine mutant.
Biochemistry, 36, 1997
|
|
1HSG
 
 | |
1HSH
 
 | |
1HSI
 
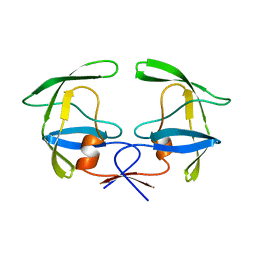 | |
1HSJ
 
 | | SARR MBP FUSION STRUCTURE | | Descriptor: | FUSION PROTEIN CONSISTING OF STAPHYLOCOCCUS ACCESSORY REGULATOR PROTEIN R AND MALTOSE BINDING PROTEIN, alpha-D-glucopyranose | | Authors: | Zhang, G. | | Deposit date: | 2000-12-26 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the SarR protein from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HSK
 
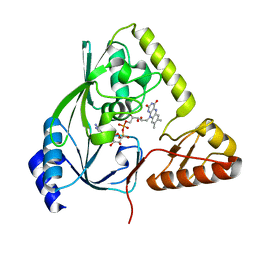 | | CRYSTAL STRUCTURE OF S. AUREUS MURB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-ACETYLENOLPYRUVOYLGLUCOSAMINE REDUCTASE | | Authors: | Benson, T.E, Harris, M.S, Choi, G.H, Cialdella, J.I, Herberg, J.T, Martin Jr, J.P, Baldwin, E.T. | | Deposit date: | 2000-12-27 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural variation for MurB: X-ray crystal structure of Staphylococcus aureus UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
Biochemistry, 40, 2001
|
|
1HSL
 
 | |
