8W0B
 
 | |
8W04
 
 | |
8W01
 
 | |
8VZR
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-bromo-o-cresol | | Descriptor: | 4-bromo-2-methylphenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VZ9
 
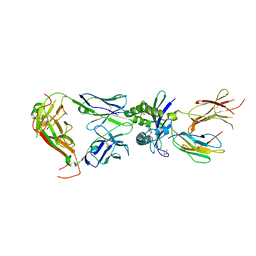 | | Crystal structure of mouse MAIT M2A TCR-MR1-5-OP-RU complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Ciacchi, L, Rossjohn, J, Awad, W. | | Deposit date: | 2024-02-11 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
J.Biol.Chem., 300, 2024
|
|
8VZ8
 
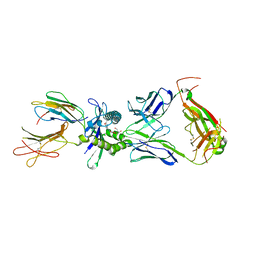 | | Crystal structure of mouse MAIT M2B TCR-MR1-5-OP-RU complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Ciacchi, L, Rossjohn, J, Awad, W. | | Deposit date: | 2024-02-11 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
J.Biol.Chem., 300, 2024
|
|
8VY4
 
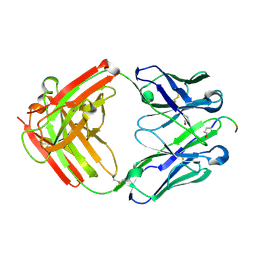 | | Engineering a Tumor-Selective Prodrug T Cell Engager Bispecific Antibody for Safer Immunotherapy | | Descriptor: | Fab Heavy Chain, Fab Light Chain, PCA-ASP-GLY-ASN-GLU-GLU-MET | | Authors: | Antonysamy, S, Demarest, S, Froning, K, Hickey, M.H, Kuhlman, B, McCue, A.C. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering a tumor-selective prodrug T-cell engager bispecific antibody for safer immunotherapy.
Mabs, 16, 2024
|
|
8VWW
 
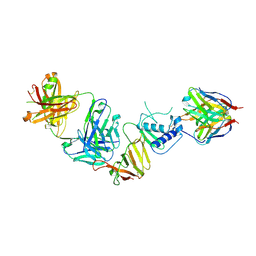 | | CCHFV GP38 bound to ADI-46152 and ADI-58048 Fabs | | Descriptor: | ADI-46152 Fab Heavy Chain, ADI-46152 Fab Light Chain, ADI-58048 Fab Heavy Chain, ... | | Authors: | Hjorth, C.K, McLellan, J.S. | | Deposit date: | 2024-02-02 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Crimean-Congo hemorrhagic fever survivors elicit protective non-neutralizing antibodies that target 11 overlapping regions on glycoprotein GP38.
Cell Rep, 43, 2024
|
|
8VW4
 
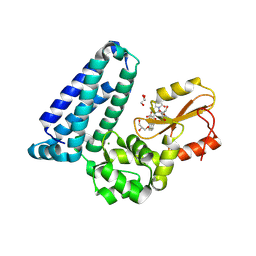 | | Crystal structure of Cbl-b TKB bound to compound 26 | | Descriptor: | (7-methoxy-2-{2-[(1S,3S,4S)-3-(3-methoxy-2-methyl-5-nitrophenyl)-1-methyl-5-oxo-1,5-dihydroimidazo[1,5-a]pyridin-2(3H)-yl]-2-oxoethoxy}quinolin-8-yl)acetic acid, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Yu, C, Murray, J, Hsu, P.L. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
8VW2
 
 | |
8VW1
 
 | |
8VW0
 
 | |
8VVL
 
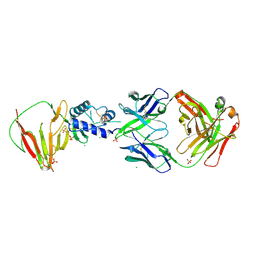 | | CCHFV GP38 bound to c13G8 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hjorth, C.K, Mishra, A.K, McLellan, J.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crimean-Congo hemorrhagic fever survivors elicit protective non-neutralizing antibodies that target 11 overlapping regions on glycoprotein GP38.
Cell Rep, 43, 2024
|
|
8VVK
 
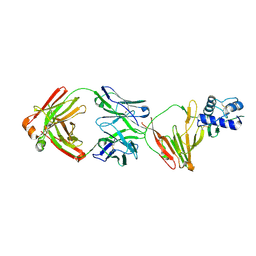 | | CCHFV GP38 bound to ADI-46143 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-46143 Fab Heavy Chain, ADI-46143 Fab Light Chain, ... | | Authors: | Hjorth, C.K, McLellan, J.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crimean-Congo hemorrhagic fever survivors elicit protective non-neutralizing antibodies that target 11 overlapping regions on glycoprotein GP38.
Cell Rep, 43, 2024
|
|
8VTT
 
 | |
8VTS
 
 | |
8VTO
 
 | |
8VTN
 
 | |
8VTL
 
 | |
8VSY
 
 | | Bile salt hydrolase from Arthrobacter citreus with covalent inhibitor AAA-10 bound | | Descriptor: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Bile salt hydrolase, CHLORIDE ION, ... | | Authors: | Dhindwal, P, Ruzzini, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bile salt hydrolase from Arthrobacter citreus with covalent inhibitor AAA-10 bound
To Be Published
|
|
8VSK
 
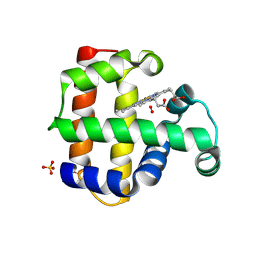 | | Crystal structure of Dehaloperoxidase A in complex with substrate 2,4-dibromophenol | | Descriptor: | 2,4-bis(bromanyl)phenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VSH
 
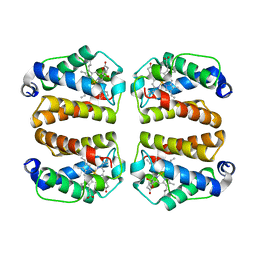 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S variant with trans heme D | | Descriptor: | Group 1 truncated hemoglobin, {3-[(2R,5'R)-9',14'-diethenyl-5'-hydroxy-5',10',15',19'-tetramethyl-5-oxo-4,5-dihydro-3H-spiro[furan-2,4'-[21,22,23,24]tetraazapentacyclo[16.2.1.13,6.18,11.113,16]tetracosa[1,3(24),6,8,10,12,14,16(22),17,19]decaen]-20'-yl-kappa~4~N~21'~,N~22'~,N~23'~,N~24'~]propanoato}iron | | Authors: | Lecomte, J.T.J, Martinez, J.E, Schlessman, J.L, Schultz, T.D, Siegler, M.A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Heme d formation in a Shewanella benthica hemoglobin.
J.Inorg.Biochem., 259, 2024
|
|
8VSG
 
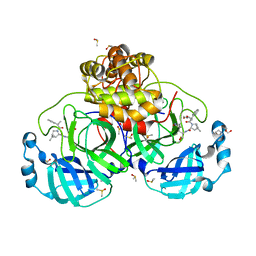 | | SARS-CoV-2 main protease with covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-(1-phenylcyclopropane-1-carbonyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Bell, J.A, Bandera, A.M. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity.
Bioorg.Med.Chem., 103, 2024
|
|
8VRX
 
 | |
8VRV
 
 | | GII.4c H2-tri HBGA norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, FORMIC ACID, ... | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2024-01-22 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Advancement of norovirus therapies
To Be Published
|
|
