4N7P
 
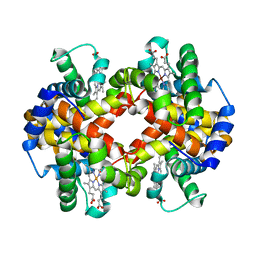 | | Capturing the haemoglobin allosteric transition in a single crystal form; Crystal structure of half-liganded human haemoglobin without phosphate at 2.8 A resolution. | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sugiyama, K, Shibayama, N, Park, S.Y. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Capturing the hemoglobin allosteric transition in a single crystal form
J.Am.Chem.Soc., 136, 2014
|
|
8HTR
 
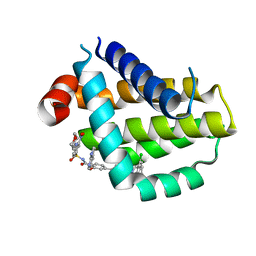 | | Crystal structure of Bcl2 in complex with S-9c | | Descriptor: | 4-[4-[(2~{S})-2-(2-chlorophenyl)pyrrolidin-1-yl]phenyl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
8HTS
 
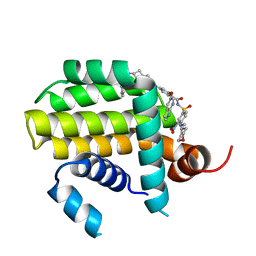 | | Crystal structure of Bcl2 in complex with S-10r | | Descriptor: | 4-[2-[(2~{S})-2-(2-cyclopropylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
4YS1
 
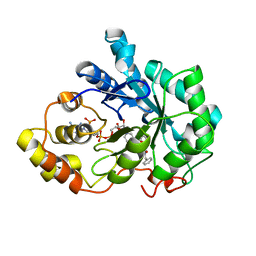 | | Human Aldose Reductase complexed with a ligand with an IDD structure (2) at 1.07 A. | | Descriptor: | 3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Price for Opening the Transient Specificity Pocket in Human Aldose Reductase upon Ligand Binding: Structural, Thermodynamic, Kinetic, and Computational Analysis.
ACS Chem. Biol., 12, 2017
|
|
8I5I
 
 | |
8I5H
 
 | |
8I0Q
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 (Local refine) | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-11 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
4YAR
 
 | | 2-Hydroxyethylphosphonate dioxygenase (HEPD) E176H | | Descriptor: | 2-hydroxyethylphosphonate dioxygenase, ACETATE ION, CADMIUM ION, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2015-02-17 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Common Late-Stage Intermediate in Catalysis by 2-Hydroxyethyl-phosphonate Dioxygenase and Methylphosphonate Synthase.
J.Am.Chem.Soc., 137, 2015
|
|
4XHZ
 
 | |
4XEN
 
 | | High pressure protein crystallography of hen egg white lysozyme in complex with Tetra-N-acetylchitotetraose at 920 MPa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Yamada, H, Watanabe, N, Nagae, T. | | Deposit date: | 2014-12-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-pressure protein crystallography of hen egg-white lysozyme
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Z1M
 
 | | Bovine F1-ATPase inhibited by three copies of the inhibitor protein IF1 crystallised in the presence of thiophosphate. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | How release of phosphate from mammalian F1-ATPase generates a rotary substep.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1JTN
 
 | |
4OTJ
 
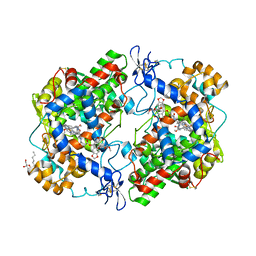 | | The complex of murine cyclooxygenase-2 with a conjugate of indomefathin and podophyllotoxin, N-{(succinylpodophyllotoxinyl)but-4-yl}-2-{1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl}acetamide | | Descriptor: | (5S,5aS,8aS,9S)-8-oxo-9-(3,4,5-trimethoxyphenyl)-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol-5-yl 4-{[4-({[1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl]acetyl}amino)butyl]amino}-4-oxobutanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-02-13 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Antitumor Activity of Cytotoxic Cyclooxygenase-2 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
1QZV
 
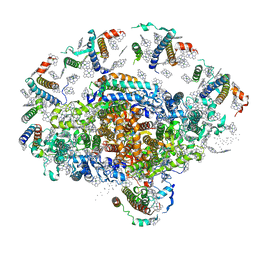 | | Crystal structure of plant photosystem I | | Descriptor: | CHLOROPHYLL A, IRON/SULFUR CLUSTER, PHYLLOQUINONE, ... | | Authors: | Ben-Shem, A, Frolow, F, Nelson, N. | | Deposit date: | 2003-09-18 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.44 Å) | | Cite: | Crystal structure of plant photosystem I.
Nature, 426, 2003
|
|
6SS0
 
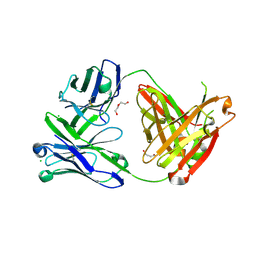 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
5L34
 
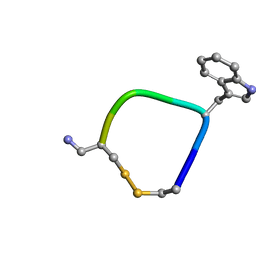 | |
6SRV
 
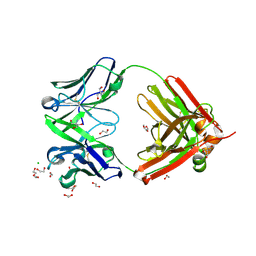 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021144 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
8AS8
 
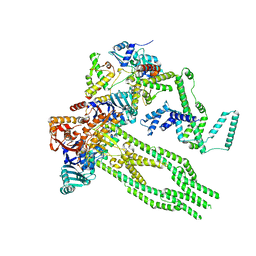 | | E. coli Wadjet JetABC monomer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, JetA, JetB, ... | | Authors: | Roisne-Hamelin, F, Beckert, B, Li, Y, Myasnikov, A, Gruber, S. | | Deposit date: | 2022-08-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage.
Mol.Cell, 82, 2022
|
|
129L
 
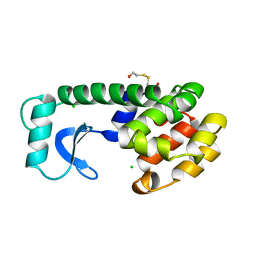 | |
130L
 
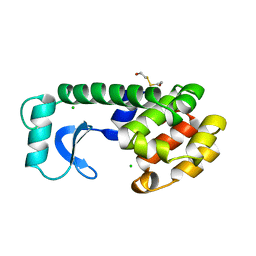 | |
131L
 
 | |
5AP3
 
 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 9-CYCLOPENTYL-2-[[2-METHOXY-4-[(1-METHYLPIPERIDIN-4-YL)OXY]-PHENYL]AMINO]-7-METHYL-7,9-DIHYDRO-8H-PURIN-8-ONE, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
3NWF
 
 | | Glycoprotein B from Herpes simplex virus type 1, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, MESO-ERYTHRITOL, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3NW8
 
 | | Glycoprotein B from Herpes simplex virus type 1, Y179S mutant, high-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75915 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3NRK
 
 | | The crystal structure of the leptospiral hypothetical protein LIC12922 | | Descriptor: | LIC12922 | | Authors: | Giuseppe, P.O, Atzingen, M.V, Nascimento, A.L.T.O, Zanchin, N.I.T, Guimaraes, B.G. | | Deposit date: | 2010-06-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of the leptospiral hypothetical protein LIC12922 reveals homology with the periplasmic chaperone SurA.
J.Struct.Biol., 173, 2011
|
|
