5AVT
 
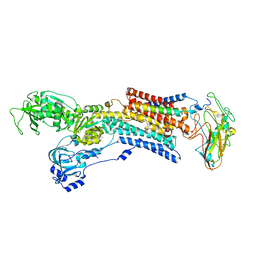 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW3
 
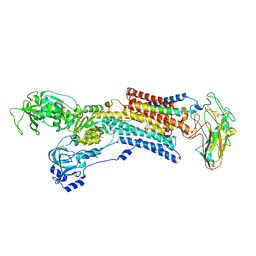 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 100 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
3LGO
 
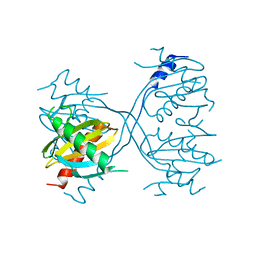 | | Structure of Gse1p, member of the GSE/EGO complex | | Descriptor: | Protein SLM4 | | Authors: | Kogan, K, Fass, D. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals.
J.Mol.Biol., 402, 2010
|
|
5AVR
 
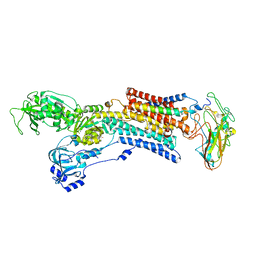 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 1.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AVZ
 
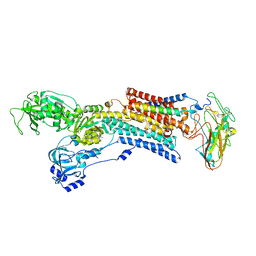 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 55 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AVU
 
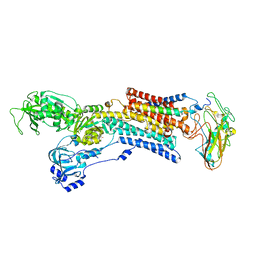 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 7.0 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW2
 
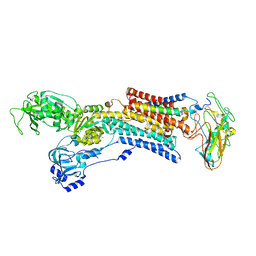 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 85 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5B2M
 
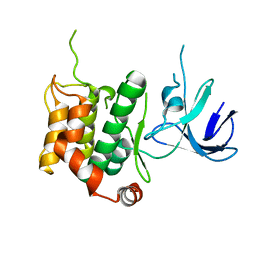 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
4N0U
 
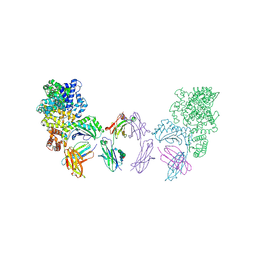 | | Ternary complex between Neonatal Fc receptor, serum albumin and Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Ig gamma-1 chain C region, ... | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2013-10-02 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Insights into Neonatal Fc Receptor-based Recycling Mechanisms.
J.Biol.Chem., 289, 2014
|
|
2ETH
 
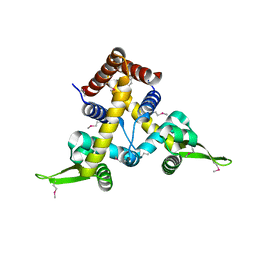 | |
4N10
 
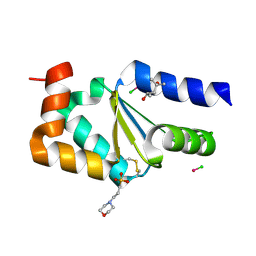 | |
4N11
 
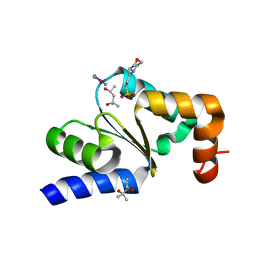 | |
2AVN
 
 | |
2EWR
 
 | |
4MZB
 
 | | Crystal structure of Grx1 from Plasmodium falciparum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Glutaredoxin | | Authors: | Yogavel, M, Sharma, A. | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.038 Å) | | Cite: | Atomic resolution crystal structure of glutaredoxin 1 from Plasmodium falciparum and comparison with other glutaredoxins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2F4P
 
 | |
4P17
 
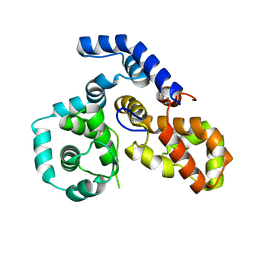 | |
2ETV
 
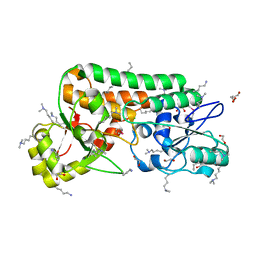 | |
1IY8
 
 | | Crystal Structure of Levodione Reductase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, LEVODIONE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sogabe, S, Fukami, T, Shiratori, Y, Yoshizumi, A, Wada, M. | | Deposit date: | 2002-07-25 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure and Stereospecificity of Levodione Reductase from Corynebacterium aquaticum M-13
J.BIOL.CHEM., 278, 2003
|
|
2F4L
 
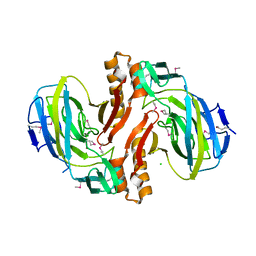 | |
2FCL
 
 | |
2JZ6
 
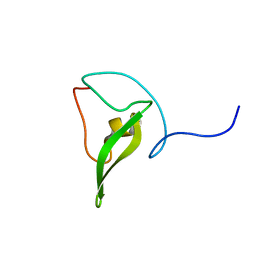 | | Solution structure of 50S ribosomal protein L28 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR97 | | Descriptor: | 50S ribosomal protein L28 | | Authors: | Wu, Y, Singarapu, K.K, Guido, V, Yee, A, Sukumaran, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 50S ribosomal protein L28 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR97.
To be Published
|
|
3QZB
 
 | |
6A6S
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans in complex with FSA, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
6A6U
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with R61G mutation, in complex with FSA | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
