1GCL
 
 | | GCN4 LEUCINE ZIPPER CORE MUTANT P-LI | | Descriptor: | GCN4 | | Authors: | Harbury, P.B, Zhang, T, Kim, P.S, Alber, T. | | Deposit date: | 1993-10-20 | | Release date: | 1995-06-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants.
Science, 262, 1993
|
|
1GCM
 
 | |
1GCN
 
 | |
1GCO
 
 | | CRYSTAL STRUCTURE OF GLUCOSE DEHYDROGENASE COMPLEXED WITH NAD+ | | Descriptor: | GLUCOSE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glucose dehydrogenase from Bacillus megaterium IWG3 at 1.7 A resolution.
J.Biochem., 129, 2001
|
|
1GCP
 
 | | CRYSTAL STRUCTURE OF VAV SH3 DOMAIN | | Descriptor: | VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
1GCQ
 
 | | CRYSTAL STRUCTURE OF VAV AND GRB2 SH3 DOMAINS | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
1GCS
 
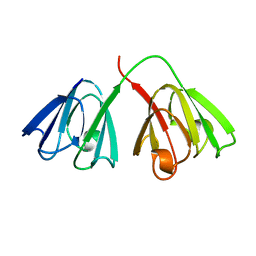 | | STRUCTURE OF THE BOVINE GAMMA-B CRYSTALLIN AT 150K | | Descriptor: | GAMMA-B CRYSTALLIN | | Authors: | Najmudin, S, Lindley, P, Slingsby, C, Bateman, O, Myles, D, Kumaraswamy, S, Glover, I. | | Deposit date: | 1994-01-27 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Bovine Gamma-B Crystallin at 150K
J.CHEM.SOC.,FARADAY TRANS., 89, 1993
|
|
1GCT
 
 | |
1GCU
 
 | |
1GCV
 
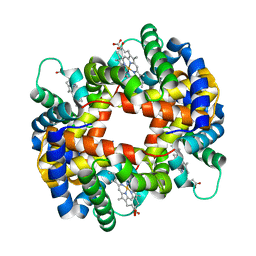 | | DEOXY FORM HEMOGLOBIN FROM MUSTELUS GRISEUS | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Naoi, Y, Chong, K.T, Yoshimatsu, K, Miyazaki, G, Tame, J.R.H, Park, S.Y, Adachi, S.I, Morimoto, H. | | Deposit date: | 2000-08-08 | | Release date: | 2000-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The functional similarity and structural diversity of human and cartilaginous fish hemoglobins.
J.Mol.Biol., 307, 2001
|
|
1GCW
 
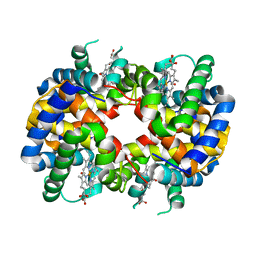 | | CO form hemoglobin from mustelus griseus | | Descriptor: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Naoi, Y, Chong, K.T, Yoshimatsu, K, Miyazaki, G, Tame, J.R.H, Park, S.Y, Adachi, S.I, Morimoto, H. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The functional similarity and structural diversity of human and cartilaginous fish hemoglobins.
J.Mol.Biol., 307, 2001
|
|
1GCY
 
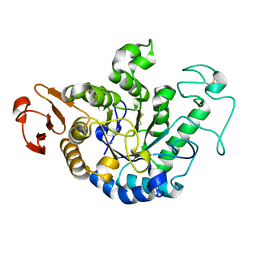 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF MALTOTETRAOSE-FORMING EXO-AMYLASE | | Descriptor: | CALCIUM ION, GLUCAN 1,4-ALPHA-MALTOTETRAHYDROLASE | | Authors: | Mezaki, Y, Katsuya, Y, Kubota, M, Matsuura, Y. | | Deposit date: | 2000-08-14 | | Release date: | 2000-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization and structural analysis of intact maltotetraose-forming exo-amylase from Pseudomonas stutzeri.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1GCZ
 
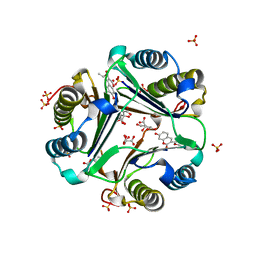 | | MACROPHAGE MIGRATION INHIBITORY FACTOR (MIF) COMPLEXED WITH INHIBITOR. | | Descriptor: | 7-HYDROXY-2-OXO-CHROMENE-3-CARBOXYLIC ACID ETHYL ESTER, CITRIC ACID, MACROPHAGE MIGRATION INHIBITORY FACTOR, ... | | Authors: | Katayama, N, Kurihara, H. | | Deposit date: | 2000-08-24 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coumarin and chromen-4-one analogues as tautomerase inhibitors of macrophage migration inhibitory factor: discovery and X-ray crystallography.
J.Med.Chem., 44, 2001
|
|
1GD0
 
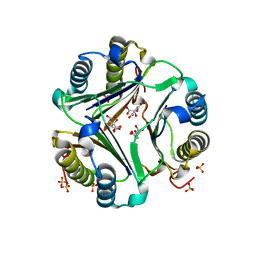 | | HUMAN MACROPHAGE MIGRATION INHIBITORY FACTOR (MIF) | | Descriptor: | CITRIC ACID, MACROPHAGE MIGRATION INHIBITORY FACTOR, SULFATE ION | | Authors: | Kurihara, H, Katayama, N. | | Deposit date: | 2000-08-24 | | Release date: | 2001-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Coumarin and chromen-4-one analogues as tautomerase inhibitors of macrophage migration inhibitory factor: discovery and X-ray crystallography.
J.Med.Chem., 44, 2001
|
|
1GD1
 
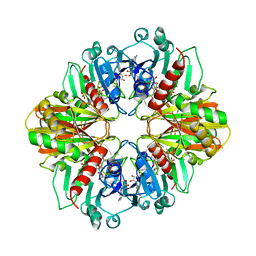 | |
1GD2
 
 | | CRYSTAL STRUCTURE OF BZIP TRANSCRIPTION FACTOR PAP1 BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*TP*AP*CP*GP*TP*AP*AP*CP*C)-3'), TRANSCRIPTION FACTOR PAP1 | | Authors: | Fujii, Y, Shimizu, T, Toda, T, Yanagida, M, Hakoshima, T. | | Deposit date: | 2000-08-25 | | Release date: | 2000-10-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the diversity of DNA recognition by bZIP transcription factors.
Nat.Struct.Biol., 7, 2000
|
|
1GD3
 
 | | refined solution structure of human cystatin A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix
conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
1GD4
 
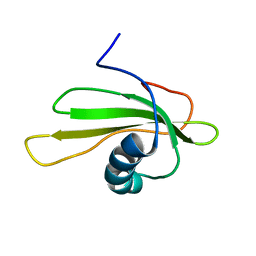 | | SOLUTION STRUCTURE OF P25S CYSTATIN A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
1GD5
 
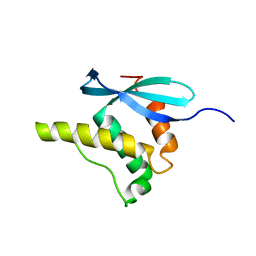 | | SOLUTION STRUCTURE OF THE PX DOMAIN FROM HUMAN P47PHOX NADPH OXIDASE | | Descriptor: | NEUTROPHIL CYTOSOL FACTOR 1 | | Authors: | Hiroaki, H, Ago, T, Ito, T, Sumimoto, H, Kohda, D. | | Deposit date: | 2000-09-14 | | Release date: | 2001-06-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PX domain, a target of the SH3 domain.
Nat.Struct.Biol., 8, 2001
|
|
1GD6
 
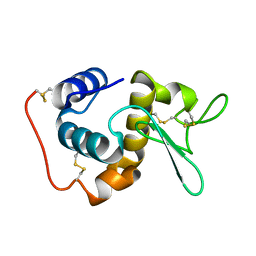 | | STRUCTURE OF THE BOMBYX MORI LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Matsuura, A, Aizawa, T, Yao, M, Kawano, K, Tanaka, I, Nitta, K. | | Deposit date: | 2000-09-19 | | Release date: | 2001-03-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of an insect lysozyme exhibiting catalytic efficiency at low temperatures.
Biochemistry, 41, 2002
|
|
1GD7
 
 | | CRYSTAL STRUCTURE OF A BIFUNCTIONAL PROTEIN (CSAA) WITH EXPORT-RELATED CHAPERONE AND TRNA-BINDING ACTIVITIES. | | Descriptor: | CSAA PROTEIN | | Authors: | Shibata, T, Inoue, Y, Vassylyev, D.G, Kawaguchi, S, Yokoyama, S, Muller, J, Linde, D, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the ttCsaA protein: an export-related chaperone from Thermus thermophilus.
EMBO J., 20, 2001
|
|
1GD8
 
 | |
1GD9
 
 | | CRYSTALL STRUCTURE OF PYROCOCCUS PROTEIN-A1 | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
1GDC
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | Deposit date: | 1994-03-15 | | Release date: | 1994-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
1GDD
 
 | |
