2OPK
 
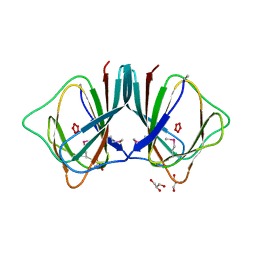 | |
1ZWX
 
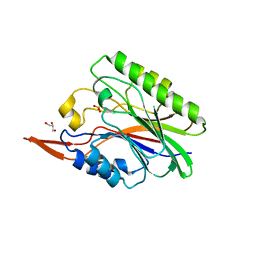 | | Crystal Structure of SmcL | | Descriptor: | GLYCEROL, PHOSPHATE ION, sphingomyelinase-c | | Authors: | Openshaw, A.E.A, Race, P.R, Monzo, H.J, Vasquez-Boland, J.A, Banfield, M.J. | | Deposit date: | 2005-06-06 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SmcL, a bacterial neutral sphingomyelinase C from Listeria.
J.Biol.Chem., 280, 2005
|
|
3P69
 
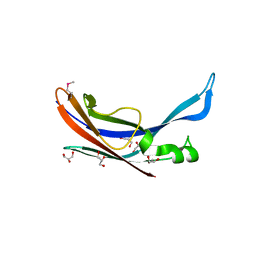 | |
2P8J
 
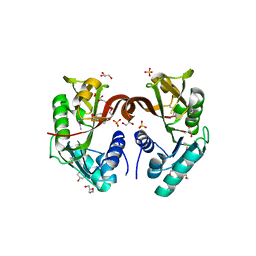 | |
3P6L
 
 | |
3P9K
 
 | |
3OW9
 
 | |
2AA5
 
 | | Mineralocorticoid Receptor with Bound Progesterone | | Descriptor: | Mineralocorticoid receptor, PROGESTERONE | | Authors: | Bledsoe, R.K, Madauss, K.P, Holt, J.A, Apolito, C.J, Lambert, M.H, Pearce, K.H, Stanley, T.B, Stewart, E.L, Trump, R.P, Willson, T.M, Williams, S.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Ligand-mediated Hydrogen Bond Network Required for the Activation of the Mineralocorticoid Receptor
J.Biol.Chem., 280, 2005
|
|
3OOX
 
 | |
3OQB
 
 | | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110 | | Descriptor: | Oxidoreductase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-02 | | Release date: | 2010-09-15 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110
To be Published
|
|
3OS7
 
 | |
3OTD
 
 | | Crystal structure of human tRNAHis guanylyltransferase (Thg1)- NaI derivative | | Descriptor: | IODIDE ION, tRNA(His) guanylyltransferase | | Authors: | Hyde, S.J, Eckenroth, B.E, Doublie, S. | | Deposit date: | 2010-09-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3P6F
 
 | |
3OTN
 
 | |
3P7W
 
 | |
1ZVC
 
 | | X-ray structure of allene oxide cyclase from arabidopsis thaliana AT3G25760 | | Descriptor: | MAGNESIUM ION, allene oxide cyclase | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | X-ray structure of allene oxide cyclase from arabidopsis thaliana AT3G25760
To be published
|
|
3P94
 
 | |
3OKO
 
 | | Crystal structure of S25-39 in complex with Kdo(2.8)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
2PR8
 
 | | crystal structure of aminoglycoside N-acetyltransferase AAC(6')-Ib11 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aminoglycoside 6-N-acetyltransferase type Ib11 | | Authors: | Maurice, F, Broutin, I, Podglajen, I, Benas, P, Collatz, E, Dardel, F. | | Deposit date: | 2007-05-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance.
Embo Rep., 9, 2008
|
|
2PRV
 
 | |
3P7S
 
 | |
3P1C
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with acetylated lysine | | Descriptor: | CREB-binding protein, N(6)-ACETYLLYSINE, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Feletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2PV4
 
 | |
3P9S
 
 | | Structure of I274A variant of E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3P6C
 
 | |
