1EEA
 
 | | Acetylcholinesterase | | Descriptor: | PROTEIN (ACETYLCHOLINESTERASE) | | Authors: | Raves, M.L, Giles, K, Schrag, J.D, Schmid, M.F, Phillips Jr, G.N, Wah, C, Howard, A.J, Silman, I, Sussman, J.L. | | Deposit date: | 1999-01-26 | | Release date: | 1999-02-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary Structure of Tetrameric Acetylcholinesterase
Structure and Function of Cholinesterases and Related Proteins, 1998
|
|
2BJ2
 
 | |
1B8C
 
 | | PARVALBUMIN | | Descriptor: | MAGNESIUM ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E.L, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-01-29 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
1B7Z
 
 | |
1B7S
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
1B5Z
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | LYSOZYME | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1BB1
 
 | | CRYSTAL STRUCTURE OF A DESIGNED, THERMOSTABLE HETEROTRIMERIC COILED COIL | | Descriptor: | CHLORIDE ION, DESIGNED, THERMOSTABLE HETEROTRIMERIC COILED COIL | | Authors: | Nautiyal, S, Alber, T. | | Deposit date: | 1998-04-28 | | Release date: | 1999-02-02 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a designed, thermostable, heterotrimeric coiled coil.
Protein Sci., 8, 1999
|
|
1F58
 
 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 24-RESIDUE PEPTIDE (RESIDUES 308-333 OF HIV-1 GP120 (MN ISOLATE) WITH ALA TO AIB SUBSTITUTION AT POSITION 323 | | Descriptor: | Envelope glycoprotein gp120, PROTEIN (IGG1 ANTIBODY 58.2 (HEAVY CHAIN)), PROTEIN (IGG1 ANTIBODY 58.2 (LIGHT CHAIN)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-21 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
1BKO
 
 | |
8CHO
 
 | |
1B7U
 
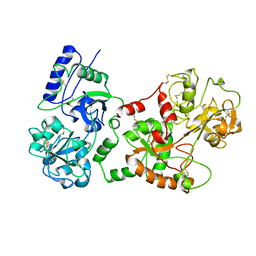 | |
1TTD
 
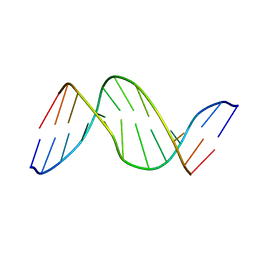 | | SOLUTION-STATE STRUCTURE OF A DNA DODECAMER DUPLEX CONTAINING A CIS-SYN THYMINE CYCLOBUTANE DIMER | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*AP*TP*TP*CP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*GP*AP*AP*(TTD)P*AP*AP*G)-3') | | Authors: | Mcateer, K, Jing, Y, Kao, J, Taylor, J.-S, Kennedy, M.A. | | Deposit date: | 1999-01-20 | | Release date: | 1999-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure of a DNA dodecamer duplex containing a Cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA.
J.Mol.Biol., 282, 1998
|
|
1BKP
 
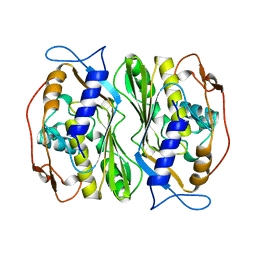 | |
1B7Q
 
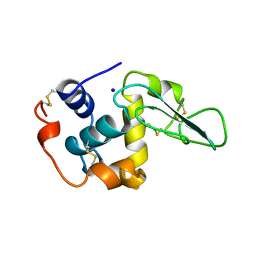 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
1B8A
 
 | | ASPARTYL-TRNA SYNTHETASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ASPARTYL-TRNA SYNTHETASE) | | Authors: | Schmitt, E, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-01-27 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation.
EMBO J., 17, 1998
|
|
1RL6
 
 | | RIBOSOMAL PROTEIN L6 | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L6) | | Authors: | Golden, B.L, Davies, C, Ramakrishnan, V, White, S.W. | | Deposit date: | 1999-01-14 | | Release date: | 1999-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ribosomal protein L6: structural evidence of gene duplication from a primitive RNA binding protein.
EMBO J., 12, 1993
|
|
2BCE
 
 | | CHOLESTEROL ESTERASE FROM BOS TAURUS | | Descriptor: | CHOLESTEROL ESTERASE | | Authors: | Chen, J.C.-H, Miercke, L.J.W, Krucinski, J, Starr, J.R, Saenz, G, Wang, X, Spilburg, C.A, Lange, L.G, Ellsworth, J.L, Stroud, R.M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic cholesterol esterase at 1.6 A: novel structural features involved in lipase activation.
Biochemistry, 37, 1998
|
|
1ZPD
 
 | | PYRUVATE DECARBOXYLASE FROM ZYMOMONAS MOBILIS | | Descriptor: | CITRIC ACID, MAGNESIUM ION, MONO-{4-[(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-AMINO]-2-HYDROXY-3-MERCAPTO-PENT-3-ENYL-PHOSPHONO} ESTER, ... | | Authors: | Lu, G, Dobritzsch, D, Schneider, G. | | Deposit date: | 1998-04-17 | | Release date: | 1999-02-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. Implications for substrate activation in pyruvate decarboxylases.
J.Biol.Chem., 273, 1998
|
|
2AY3
 
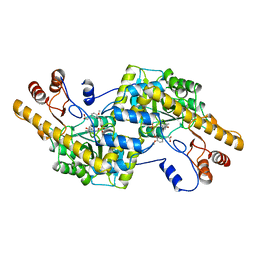 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITH 3-(3,4-DIMETHOXYPHENYL)PROPIONIC ACID | | Descriptor: | 3-(3,4-DIMETHOXYPHENYL)PROPIONIC ACID, AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1998-08-06 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active site of Paracoccus denitrificans aromatic amino acid aminotransferase has contrary properties: flexibility and rigidity.
Biochemistry, 38, 1999
|
|
2AY9
 
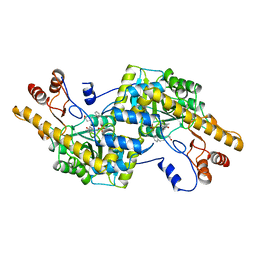 | |
1BJ2
 
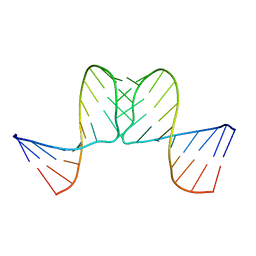 | |
1BJ6
 
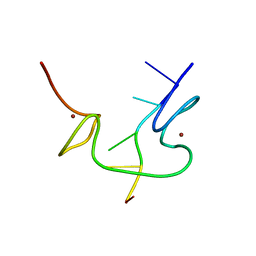 | | 1H NMR OF (12-53) NCP7/D(ACGCC) COMPLEX, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), NUCLEOCAPSID PROTEIN 7, ZINC ION | | Authors: | Demene, H, Morellet, N, Teilleux, V, Huynh-Dinh, T, De Rocquigny, H, Fournie-Zaluski, M.C, Roques, B.P. | | Deposit date: | 1998-07-03 | | Release date: | 1999-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC).
J.Mol.Biol., 283, 1998
|
|
2AY6
 
 | |
2AY4
 
 | |
2AY8
 
 | |
