2FIB
 
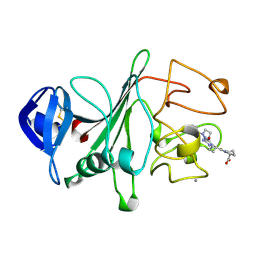 | | RECOMBINANT HUMAN GAMMA-FIBRINOGEN CARBOXYL TERMINAL FRAGMENT (RESIDUES 143-411) COMPLEXED TO THE PEPTIDE GLY-PRO-ARG-PRO AT PH 6.0 | | Descriptor: | CALCIUM ION, FIBRINOGEN, GLY-PRO-ARG-PRO | | Authors: | Pratt, K.P, Cote, H.C.F, Chung, D.W, Stenkamp, R.E, Davie, E.W. | | Deposit date: | 1997-06-03 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal gamma chain fragment complexed with the peptide Gly-Pro-Arg-Pro.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1TOH
 
 | |
1DSS
 
 | |
1ALW
 
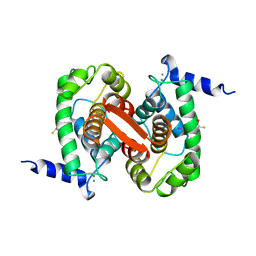 | | INHIBITOR AND CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | Descriptor: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, CALPAIN | | Authors: | Narayana, S.V.L, Lin, G. | | Deposit date: | 1997-06-04 | | Release date: | 1998-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
3TMY
 
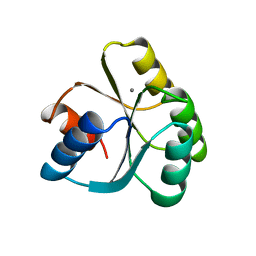 | | CHEY FROM THERMOTOGA MARITIMA (MN-III) | | Descriptor: | CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-04 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
1GW3
 
 | |
1GW4
 
 | |
1JSA
 
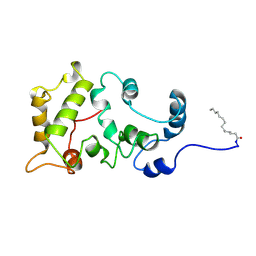 | | MYRISTOYLATED RECOVERIN WITH TWO CALCIUMS BOUND, NMR, 24 STRUCTURES | | Descriptor: | CALCIUM ION, MYRISTIC ACID, RECOVERIN | | Authors: | Ames, J.B, Ishima, R, Tanaka, T, Gordon, J.I, Stryer, L, Ikura, M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanics of calcium-myristoyl switches.
Nature, 389, 1997
|
|
2EZD
 
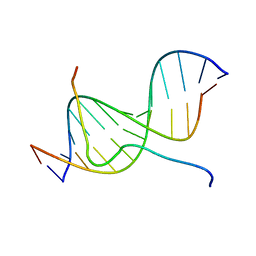 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZE
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZF
 
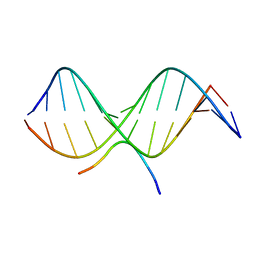 | | SOLUTION STRUCTURE OF A COMPLEX OF THE THIRD DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZG
 
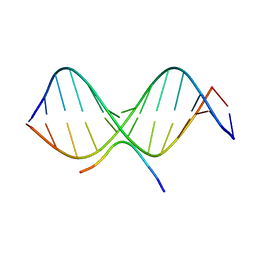 | | SOLUTION STRUCTURE OF A COMPLEX OF THE THIRD DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
1ALY
 
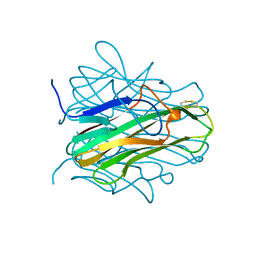 | |
335D
 
 | |
1FSP
 
 | | NMR SOLUTION STRUCTURE OF BACILLUS SUBTILIS SPO0F PROTEIN, 20 STRUCTURES | | Descriptor: | STAGE 0 SPORULATION PROTEIN F | | Authors: | Feher, V.A, Skelton, N.J, Dahlquist, F.W, Cavanagh, J. | | Deposit date: | 1997-06-05 | | Release date: | 1997-12-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Biochemistry, 36, 1997
|
|
1ALX
 
 | | GRAMICIDIN D FROM BACILLUS BREVIS (METHANOL SOLVATE) | | Descriptor: | GRAMICIDIN A, METHANOL | | Authors: | Burkhart, B.M, Langs, D.A, Smith, G.D, Courseille, C, Precigoux, G, Hospital, M, Pangborn, W.A, Duax, W.L. | | Deposit date: | 1997-06-05 | | Release date: | 1998-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Heterodimer Formation and Crystal Nucleation of Gramicidin D
Biophys.J., 75, 1998
|
|
1AL0
 
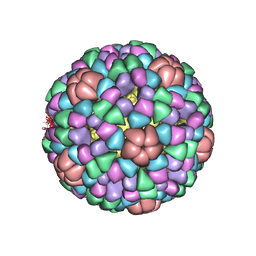 | | PROCAPSID OF BACTERIOPHAGE PHIX174 | | Descriptor: | CAPSID PROTEIN GPF, SCAFFOLDING PROTEIN GPB, SCAFFOLDING PROTEIN GPD, ... | | Authors: | Rossmann, M.G, Dokland, T. | | Deposit date: | 1997-06-06 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a viral procapsid with molecular scaffolding.
Nature, 389, 1997
|
|
4TMY
 
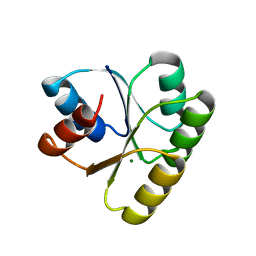 | | CHEY FROM THERMOTOGA MARITIMA (MG-IV) | | Descriptor: | CHEY PROTEIN, MAGNESIUM ION | | Authors: | Usher, K.C, De La Cruz, A, Dahlquist, F.W, Remington, S.J. | | Deposit date: | 1997-06-06 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability.
Protein Sci., 7, 1998
|
|
1ALZ
 
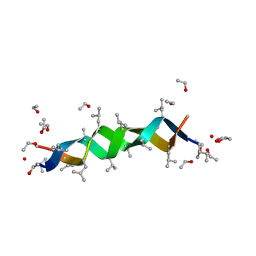 | | GRAMICIDIN D FROM BACILLUS BREVIS (ETHANOL SOLVATE) | | Descriptor: | ETHANOL, ILE-GRAMICIDIN C, VAL-GRAMICIDIN A | | Authors: | Burkhart, B.M, Pangborn, W.A, Duax, W.L, Langs, D.A. | | Deposit date: | 1997-06-06 | | Release date: | 1998-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Heterodimer Formation and Crystal Nucleation of Gramicidin D
Biophys.J., 75, 1998
|
|
1UEA
 
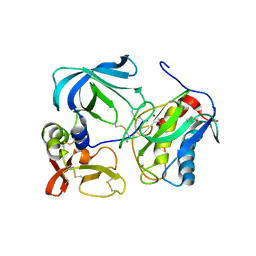 | | MMP-3/TIMP-1 COMPLEX | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-3, TISSUE INHIBITOR OF METALLOPROTEINASE-1, ... | | Authors: | Bode, W, Maskos, K, Gomis-Rueth, F.-X, Nagase, H. | | Deposit date: | 1997-06-06 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1.
Nature, 389, 1997
|
|
1XCH
 
 | |
2FSP
 
 | | NMR SOLUTION STRUCTURE OF BACILLUS SUBTILIS SPO0F PROTEIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STAGE 0 SPORULATION PROTEIN F | | Authors: | Feher, V.A, Skelton, N.J, Dahlquist, F.W, Cavanagh, J. | | Deposit date: | 1997-06-06 | | Release date: | 1997-12-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Biochemistry, 36, 1997
|
|
1JOI
 
 | | STRUCTURE OF PSEUDOMONAS FLUORESCENS HOLO AZURIN | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Lee, X, Ton-that, H, Zhu, D.-W, Biesterfedlt, J, Lanthier, P.H, Yachuchi, M, Dahms, T, Szabo, A.G. | | Deposit date: | 1997-06-09 | | Release date: | 1997-12-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallization and preliminary crystallographic studies of the crystals of the azurin Pseudomonas fluorescens.
Arch.Biochem.Biophys., 308, 1994
|
|
1FSD
 
 | |
1AL2
 
 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT V1160I | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-06-09 | | Release date: | 1997-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
