7TN4
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 3-diphosphoinositol 1,2,4,5-tetrakisphosphate (3-PP-IP4), Mg and Fluoride ion | | Descriptor: | (1R,2S,3R,4R,5S,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2022-01-20 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and catalytic analyses of the InsP 6 kinase activities of higher plant ITPKs.
Faseb J., 36, 2022
|
|
7TN8
 
 | |
7TN3
 
 | |
7TN7
 
 | |
3ZUD
 
 | | THERMOASCUS GH61 ISOZYME A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Otten, H, Quinlan, R.J, Sweeney, M.D, Poulsen, J.-C.N, Johansen, K.S, Krogh, K.B.R.M, Joergensen, C.I, Tovborg, M, Anthonsen, A, Tryfona, T, Walter, C.P, Dupree, P, Xu, F, Davies, G.J, Walton, P.H, Lo Leggio, L. | | Deposit date: | 2011-07-18 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Insights Into the Oxidative Degradation of Cellulose by a Copper Metalloenzyme that Exploits Biomass Components.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2D5F
 
 | | Crystal Structure of Recombinant Soybean Proglycinin A3B4 subunit, its Comparison with Mature Glycinin A3B4 subunit, Responsible for Hexamer Assembly | | Descriptor: | CARBONATE ION, MAGNESIUM ION, glycinin A3B4 subunit | | Authors: | Itoh, T, Adachi, M, Masuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2005-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
2VO2
 
 | | Crystal structure of soybean ascorbate peroxidase mutant W41A subjected to low dose X-rays | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Metcalfe, C.L, Badyal, S.K, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron Oxidation State Modulates Active Site Structure in a Heme Peroxidase.
Biochemistry, 47, 2008
|
|
2WSF
 
 | | Improved Model of Plant Photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3G54890, BETA-CAROTENE, ... | | Authors: | Amunts, A, Toporik, H, Borovikov, A, Nelson, N. | | Deposit date: | 2009-09-05 | | Release date: | 2009-11-17 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structure determination and improved model of plant photosystem I.
J. Biol. Chem., 285, 2010
|
|
2DQX
 
 | | mutant beta-amylase (W55R) from soy bean | | Descriptor: | Beta-amylase | | Authors: | Ishikawa, K. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and structural analysis of enzyme sliding on a substrate: multiple attack in beta-amylase
Biochemistry, 46, 2007
|
|
3A68
 
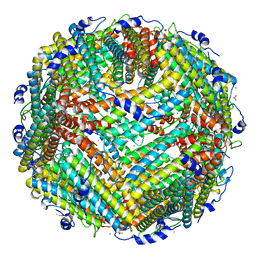 | | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin | | Descriptor: | ACETIC ACID, CALCIUM ION, Ferritin-4, ... | | Authors: | Masuda, T, Goto, F, Yoshihara, T, Mikami, B. | | Deposit date: | 2009-08-26 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin
J.Biol.Chem., 285, 2010
|
|
8FSD
 
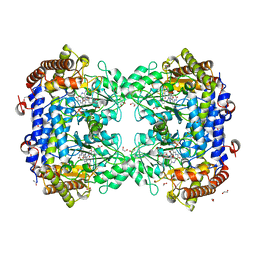 | | P130R mutant of soybean SHMT8 in complex with PLP-glycine and formylTHF | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Beamer, L.J, Korasick, D.A. | | Deposit date: | 2023-01-09 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional analysis of two SHMT8 variants associated with soybean cyst nematode resistance.
Febs J., 291, 2024
|
|
2XIF
 
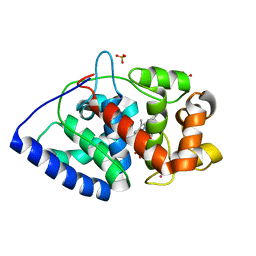 | | The structure of ascorbate peroxidase Compound II | | Descriptor: | ASCORBATE PEROXIDASE, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
8R37
 
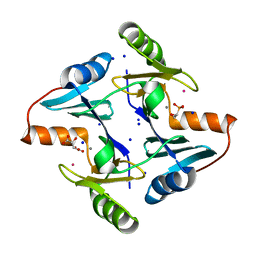 | | Klebsiella pneumoniae fosfomycin-resistance protein (FosAKP) | | Descriptor: | FOSFOMYCIN, FosA family fosfomycin resistance glutathione transferase, L(+)-TARTARIC ACID, ... | | Authors: | Papageorgiou, A.C, Varotsou, C, Labrou, N.E. | | Deposit date: | 2023-11-08 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Studies of Klebsiella pneumoniae Fosfomycin-Resistance Protein and Its Application for the Development of an Optical Biosensor for Fosfomycin Determination.
Int J Mol Sci, 25, 2023
|
|
2VNX
 
 | | Crystal structure of soybean ascorbate peroxidase mutant W41A after exposure to a high dose of x-rays | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Metcalfe, C.L, Badyal, S.K, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Iron Oxidation State Modulates Active Site Structure in a Heme Peroxidase.
Biochemistry, 47, 2008
|
|
2WSC
 
 | | Improved Model of Plant Photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3G54890, BETA-CAROTENE, ... | | Authors: | Amunts, A, Toporik, H, Borovikov, A, Nelson, N. | | Deposit date: | 2009-09-04 | | Release date: | 2009-11-17 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure Determination and Improved Model of Plant Photosystem I.
J.Biol.Chem., 285, 2010
|
|
2WSE
 
 | | Improved Model of Plant Photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3G54890, BETA-CAROTENE, ... | | Authors: | Amunts, A, Toporik, H, Borovikov, A, Nelson, N. | | Deposit date: | 2009-09-05 | | Release date: | 2009-11-17 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure determination and improved model of plant photosystem I
J. Biol. Chem., 285, 2010
|
|
2XIH
 
 | | The structure of ascorbate peroxidase Compound III | | Descriptor: | ASCORBATE PEROXIDASE, OXYGEN MOLECULE, POTASSIUM ION, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2XJ6
 
 | | The structure of ferrous ascorbate peroxidase | | Descriptor: | ASCORBATE PEROXIDASE, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2VNZ
 
 | | Crystal structure of dithinonite reduced soybean ascorbate peroxidase mutant W41A. | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Metcalfe, C.L, Badyal, S.K, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Iron Oxidation State Modulates Active Site Structure in a Heme Peroxidase.
Biochemistry, 47, 2008
|
|
2WD4
 
 | | Ascorbate Peroxidase as a heme oxygenase: w41A variant product with t-butyl peroxide | | Descriptor: | 3-[2-[[3-(2-CARBOXYETHYL)-5-[[3-ETHENYL-4-METHYL-5-[(2-METHYLPROPAN-2-YL)OXY]-1H-PYRROL-2-YL]METHYL]-4-METHYL-1H-PYRROL -2-YL]METHYL]-5-[(Z)-(4-ETHENYL-3-METHYL-5-OXO-PYRROL-2-YLIDENE)METHYL]-4-METHYL-1H-PYRROL-3-YL]PROPANOIC ACID, ASCORBATE PEROXIDASE, FE (III) ION, ... | | Authors: | Badyal, S.K, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for Heme Oxygenase Activity in a Heme Peroxidase.
Biochemistry, 48, 2009
|
|
3A9Q
 
 | | Crystal Structure Analysis of E173A variant of the soybean ferritin SFER4 | | Descriptor: | ACETIC ACID, CALCIUM ION, Ferritin-4, ... | | Authors: | Masuda, T, Goto, F, Yoshihara, T, Mikami, B. | | Deposit date: | 2009-11-05 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin
J.Biol.Chem., 285, 2010
|
|
2D5H
 
 | | Crystal Structure of Recombinant Soybean Proglycinin A3B4 subunit, its Comparison with Mature Glycinin A3B4 subunit, Responsible for Hexamer Assembly | | Descriptor: | CARBONATE ION, MAGNESIUM ION, glycinin A3B4 subunit | | Authors: | Itoh, T, Adachi, M, Masuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2005-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
5VAB
 
 | | Crystal structure of ATXR5 PHD domain in complex with histone H3 | | Descriptor: | ATXR5 PHD domain, Histone H3 peptide, ZINC ION | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5WHX
 
 | | PREPHENATE DEHYDROGENASE FROM SOYBEAN | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prephenate dehydrogenase 1 | | Authors: | Holland, C.K, Jez, J.M. | | Deposit date: | 2017-07-18 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular basis of the evolution of alternative tyrosine biosynthetic routes in plants.
Nat. Chem. Biol., 13, 2017
|
|
2SBL
 
 | |
