311D
 
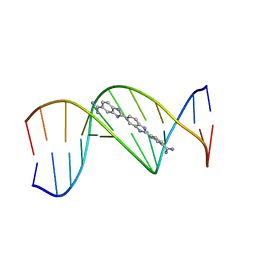 | | THE ROLE OF HYDROGEN BONDING IN MINOR-GROOVE DRUG-DNA RECOGNITION. STRUCTURE OF A BIS-AMIDINIUM DERIVATIVE OF HOECHST 33258 COMPLEXED TO THE DODECANUCLEOTIDE D(CGCGAATTCGCG)2 | | Descriptor: | 5-AMIDINO-2-[2-(4-AMIDINOPHENYL)-5-BENZIMIDAZOLYL]BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Clark, G.R, Boykin, D.W, Czarny, A, Neidle, S. | | Deposit date: | 1997-02-04 | | Release date: | 1997-02-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a bis-amidinium derivative of hoechst 33258 complexed to dodecanucleotide d(CGCGAATTCGCG)2: the role of hydrogen bonding in minor groove drug-DNA recognition.
Nucleic Acids Res., 25, 1997
|
|
314D
 
 | | Z-DNA HEXAMER WITH 5' OVERHANGS THAT FORM A REVERSE WOBBLE BASE PAIR | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Mooers, B.H.M, Eichman, B.F, Ho, P.S. | | Deposit date: | 1997-02-04 | | Release date: | 1997-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures and relative stabilities of d(G x G) reverse Hoogsteen, d(G x T) reverse wobble, and d(G x C) reverse Watson-Crick base-pairs in DNA crystals.
J.Mol.Biol., 269, 1997
|
|
1SZJ
 
 | |
1AB8
 
 | |
1AB5
 
 | | STRUCTURE OF CHEY MUTANT F14N, V21T | | Descriptor: | CHEY | | Authors: | Wilcock, D, Pisabarro, M.T, Lopez-Hernandez, E, Serrano, L, Coll, M. | | Deposit date: | 1997-02-04 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure analysis of two CheY mutants: importance of the hydrogen-bond contribution to protein stability.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AB6
 
 | | STRUCTURE OF CHEY MUTANT F14N, V86T | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Wilcock, D, Pisabarro, M.T, Lopez-Hernandez, E, Serranno, L, Coll, M. | | Deposit date: | 1997-02-04 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of two CheY mutants: importance of the hydrogen-bond contribution to protein stability.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1SMG
 
 | | CALCIUM-BOUND E41A MUTANT OF THE N-DOMAIN OF CHICKEN TROPONIN C, NMR, 40 STRUCTURES | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Gagne, S.M, Li, M.X, Sykes, B.D. | | Deposit date: | 1997-02-04 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of direct coupling between binding and induced structural change in regulatory calcium binding proteins.
Biochemistry, 36, 1997
|
|
1NCV
 
 | | DETERMINATION CC-CHEMOKINE MCP-3, NMR, 7 STRUCTURES | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 3 | | Authors: | Meunier, S, Bernassau, J.M, Guillemot, J.C, Ferrara, P, Darbon, H. | | Deposit date: | 1997-02-05 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of CC chemokine monocyte chemoattractant protein 3 by 1H two-dimensional NMR spectroscopy.
Biochemistry, 36, 1997
|
|
1AB9
 
 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN | | Descriptor: | GAMMA-CHYMOTRYPSIN, PENTAPEPTIDE (TPGVY), SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-02-05 | | Release date: | 1997-08-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
1ACC
 
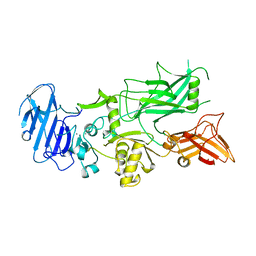 | | ANTHRAX PROTECTIVE ANTIGEN | | Descriptor: | ANTHRAX PROTECTIVE ANTIGEN, CALCIUM ION | | Authors: | Petosa, C, Liddington, R.C. | | Deposit date: | 1997-02-05 | | Release date: | 1998-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the anthrax toxin protective antigen.
Nature, 385, 1997
|
|
1TSK
 
 | |
1ACD
 
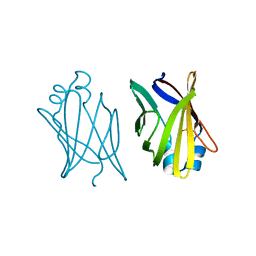 | | V32D/F57H MUTANT OF MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Kane, C.D, Simpson, M, Banaszak, L.J, Bernlohr, D.A. | | Deposit date: | 1997-02-06 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
J.Biol.Chem., 272, 1997
|
|
2CJN
 
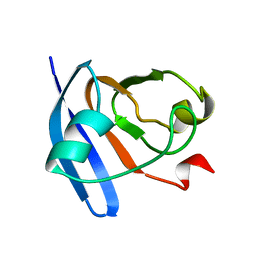 | | STRUCTURE OF FERREDOXIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FERREDOXIN | | Authors: | Hatanaka, H, Tanimura, R, Katoh, S, Inagaki, F. | | Deposit date: | 1997-02-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ferredoxin from the thermophilic cyanobacterium Synechococcus elongatus and its thermostability.
J.Mol.Biol., 268, 1997
|
|
2CJO
 
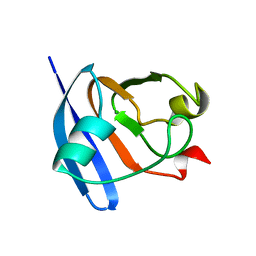 | | STRUCTURE OF FERREDOXIN, NMR, 10 STRUCTURES | | Descriptor: | FERREDOXIN | | Authors: | Hatanaka, H, Tanimura, R, Katoh, S, Inagaki, F. | | Deposit date: | 1997-02-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ferredoxin from the thermophilic cyanobacterium Synechococcus elongatus and its thermostability.
J.Mol.Biol., 268, 1997
|
|
2MPR
 
 | | MALTOPORIN FROM SALMONELLA TYPHIMURIUM | | Descriptor: | CALCIUM ION, MALTOPORIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Meyer, J.E.W, Schulz, G.E. | | Deposit date: | 1997-02-07 | | Release date: | 1997-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of maltoporin from Salmonella typhimurium ligated with a nitrophenyl-maltotrioside.
J.Mol.Biol., 266, 1997
|
|
1KSR
 
 | | THE REPEATING SEGMENTS OF THE F-ACTIN CROSS-LINKING GELATION FACTOR (ABP-120) HAVE AN IMMUNOGLOBULIN FOLD, NMR, 20 STRUCTURES | | Descriptor: | GELATION FACTOR | | Authors: | Fucini, P, Renner, C, Herberhold, C, Noegel, A.A, Holak, T.A. | | Deposit date: | 1997-02-07 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold.
Nat.Struct.Biol., 4, 1997
|
|
1ACI
 
 | |
1MJY
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D70N | | Descriptor: | INORGANIC PYROPHOSPHATASE | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1MJW
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D42N | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Samygina, V.R, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1MJX
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D65N | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1FGK
 
 | |
1MJZ
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D97N | | Descriptor: | INORGANIC PYROPHOSPHATASE | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1ACW
 
 | | SOLUTION NMR STRUCTURE OF P01, A NATURAL SCORPION PEPTIDE STRUCTURALLY ANALOGOUS TO SCORPION TOXINS SPECIFIC FOR APAMIN-SENSITIVE POTASSIUM CHANNEL, 25 STRUCTURES | | Descriptor: | NATURAL SCORPION PEPTIDE P01 | | Authors: | Blanc, E, Fremont, V, Sizun, P, Meunier, S, Van Rietschoten, J, Thevand, A, Bernassau, J.M, Darbon, H. | | Deposit date: | 1997-02-10 | | Release date: | 1997-04-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P01, a natural scorpion peptide structurally analogous to scorpion toxins specific for apamin-sensitive potassium channel.
Proteins, 24, 1996
|
|
1ACV
 
 | | DSBA MUTANT H32S | | Descriptor: | DSBA | | Authors: | Guddat, L.W, Martin, J.L. | | Deposit date: | 1997-02-10 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability.
Protein Sci., 6, 1997
|
|
1VZV
 
 | | STRUCTURE OF VARICELLA-ZOSTER VIRUS PROTEASE | | Descriptor: | VARICELLA-ZOSTER VIRUS PROTEASE | | Authors: | Qiu, X, Jason, C.A, Culp, J.S, Richardson, S.B, Debouck, C, Smith, W.W, Abdel-Meguid, S.S. | | Deposit date: | 1997-02-10 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of varicella-zoster virus protease.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
