5QM9
 
 | |
5QMO
 
 | |
5QNB
 
 | |
5QNS
 
 | |
5QO6
 
 | |
5QKP
 
 | |
5QL5
 
 | |
5QLG
 
 | |
5QLY
 
 | |
5QN8
 
 | |
5QNM
 
 | |
5QO1
 
 | |
5QOG
 
 | |
5QKN
 
 | |
5QL4
 
 | |
5QLH
 
 | |
5QLX
 
 | |
5QMD
 
 | |
5QMS
 
 | |
5QN9
 
 | |
5QNQ
 
 | |
5QO7
 
 | |
6L3X
 
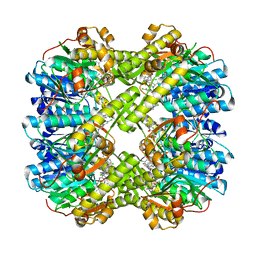 | | Discovery of novel peptidomimetic boronate ClpP inhibitors with noncanonical enzyme mechanism as potent virulence blockers in vitro and in vivo | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, [(1~{R})-1-[[(2~{S})-2-[[2,5-bis(chloranyl)phenyl]carbonylamino]-3-(1~{H}-indol-3-yl)propanoyl]amino]-3-methyl-butyl]boronic acid | | Authors: | Luo, Y.F, Bao, R, Ju, Y, He, L.H. | | Deposit date: | 2019-10-15 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3054 Å) | | Cite: | Discovery of Novel Peptidomimetic Boronate ClpP Inhibitors with Noncanonical Enzyme Mechanism as Potent Virulence Blockersin Vitroandin Vivo.
J.Med.Chem., 63, 2020
|
|
6LH7
 
 | |
5QKO
 
 | |
