8CPP
 
 | | CRYSTAL STRUCTURES OF CYTOCHROME P450-CAM COMPLEXED WITH CAMPHANE, THIOCAMPHOR, AND ADAMANTANE: FACTORS CONTROLLING P450 SUBSTRATE HYDROXYLATION | | Descriptor: | CYTOCHROME P450CAM, PROTOPORPHYRIN IX CONTAINING FE, THIOCAMPHOR | | Authors: | Raag, R, Poulos, T.L. | | Deposit date: | 1990-05-18 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of cytochrome P-450CAM complexed with camphane, thiocamphor, and adamantane: factors controlling P-450 substrate hydroxylation.
Biochemistry, 30, 1991
|
|
8CPL
 
 | |
8CPK
 
 | |
8CPD
 
 | | Cryo-EM structure of CRaf dimer with 14:3:3 | | Descriptor: | 14-3-3 protein zeta isoform X1, RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Dedden, D, Graedler, U, Schwarz, D, Thomsen, M, Leuthner, B, Schneider, E, Nitsche, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
8CPB
 
 | | 1,6-anhydro-n-actetylmuramic acid kinase (AnmK) in complex with AMPPNP, and AnhMurNAc at 1.7 Angstroms resolution. | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, GLYCEROL, ... | | Authors: | Jimenez-Faraco, E, Hermoso, J.A. | | Deposit date: | 2023-03-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic process of anhydro-N-acetylmuramic acid kinase from Pseudomonas aeruginosa.
J.Biol.Chem., 299, 2023
|
|
8CPA
 
 | |
8CP6
 
 | |
8CP5
 
 | |
8CP4
 
 | | [4Fe-4S] cluster containing LarE in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Zecchin, P, Pecqueur, L, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-03-01 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
8CP3
 
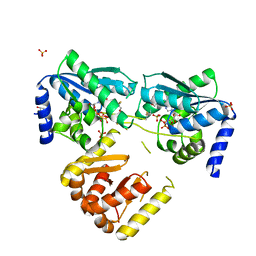 | | Apo-LarE in complex with AMP-PNP | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, NAD_synthase domain-containing protein, ... | | Authors: | Zecchin, P, Pecqueur, L, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-03-01 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
8CP2
 
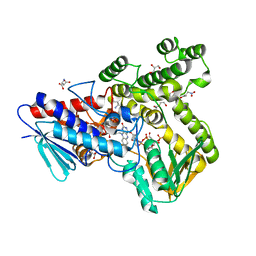 | |
8CP0
 
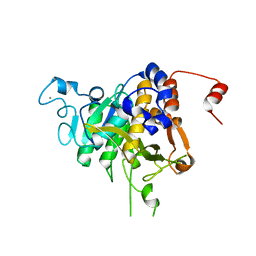 | | Structure of the catalytic domain of P. vivax Sub1 (trigonal crystal form) | | Descriptor: | CALCIUM ION, subtilisin | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8COZ
 
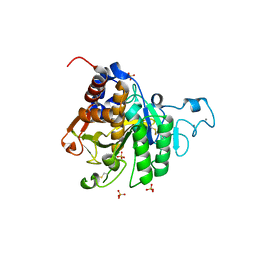 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.438 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8COY
 
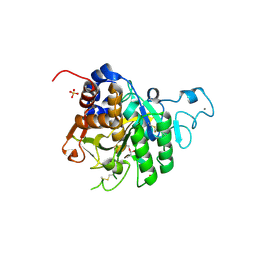 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) in complex with inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
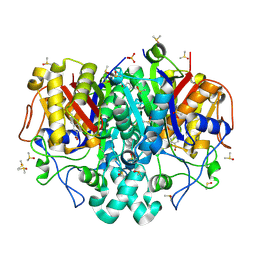
8COV
 
 | |
8COU
 
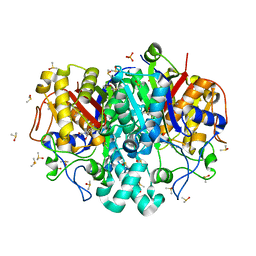 | | Pa.FabF-C164Q in complex with 3-acetamido-4-methoxybenzoic acid | | Descriptor: | 3-acetamido-4-methoxy-benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiou, C, Brenk, R. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
8COP
 
 | |
8COM
 
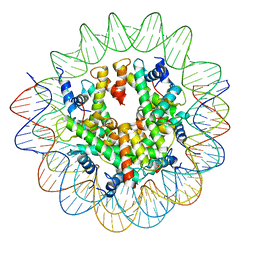 | | Structure of the Nucleosome Core Particle from Trypanosoma brucei | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Burdett, H, Deak, G, Wilson, M.D. | | Deposit date: | 2023-02-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Histone divergence in trypanosomes results in unique alterations to nucleosome structure.
Nucleic Acids Res., 51, 2023
|
|
8COK
 
 | |
8COI
 
 | | Human adenovirus-derived synthetic ADDobody binder | | Descriptor: | ADDobody | | Authors: | Buzas, D, Toelzer, C, Gupta, K, Berger-Schaffitzel, C, Berger, I. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8COH
 
 | | Structure of the complement C5 specific nanobody TPP-3444 | | Descriptor: | CITRIC ACID, MANGANESE (II) ION, Nanobody TPP-3444 | | Authors: | Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of the bispecific VHH antibody gefurulimab (ALXN1720) targeting complement component 5, and designed for low volume subcutaneous administration.
Mol.Immunol., 165, 2023
|
|
8CO5
 
 | |
8CO2
 
 | |
8CO0
 
 | |
8CNZ
 
 | | mmLarE-[4Fe-4S] phased by Fe-SAD | | Descriptor: | CHLORIDE ION, IODIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Pecqueur, L, Zecchin, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-02-24 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
