5QCT
 
 | | Crystal structure of BACE complex with BMC001 | | Descriptor: | (2R,4S)-N-butyl-4-[(4S,6R)-16-ethoxy-12-ethyl-6-methyl-2,13-dioxo-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-trien-4-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1, PHOSPHATE ION | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD4
 
 | | Crystal structure of BACE complex with BMC023 | | Descriptor: | Beta-secretase 1, {(E)-(3R,6S,9R)-3-[(1S,3R)-3-((S)-1 -BUTYLCARBAMOYL-2-METHYL-PROPYLCARB AMOYL)-1-HYDROXY-BUTYL]-6-METHYL-5, 8-DIOXO-1,11-DITHIA-4,7-DIAZA-CYCLO PENTADEC-13-EN-9-YL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
7KN5
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobodies VHH E and U | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
4ZP5
 
 | | MAP4K4 in complex with inhibitor | | Descriptor: | 4-[5-(4-chlorophenyl)-1,3-oxazol-2-yl]benzamide, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Liu, S. | | Deposit date: | 2015-05-07 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of MAP4K4 complex
To Be Published
|
|
5YSQ
 
 | | Sulfate-complex structure of a pyrophosphate-dependent kinase in the ribokinase family provides insight into the donor-binding mode | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, SULFATE ION, TM0415 | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Identification of a pyrophosphate-dependent kinase and its donor selectivity determinants.
Nat Commun, 9, 2018
|
|
4BFS
 
 | | Crystal structure of Mycobacterium tuberculosis PanK in complex with a triazole inhibitory compound (1a) | | Descriptor: | N-[1-(5-{[2-(4-FLUOROPHENOXY)ETHYL]SULFANYL}-4-METHYL-4H-1,2,4-TRIAZOL-3-YL)ETHYL]-2-(TRIFLUOROMETHYL)BENZAMIDE, PANTOTHENATE KINASE | | Authors: | Bjorkelid, C, Bergfors, T, Jones, T.A. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Biochemical Characterization of Compounds Inhibiting Mycobacterium Tuberculosis Pank
J.Biol.Chem., 288, 2013
|
|
5QF6
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMSOA000281b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-methyl-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridine-3-carbonitrile, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QE9
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000232a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloro-N-cyclopentyl-1-methyl-1H-pyrazole-3-carboxamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QEM
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_XST00000217b | | Descriptor: | 2-(thiophen-2-yl)-1,3-thiazole-4-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFN
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000324a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
3G5O
 
 | | The crystal structure of the toxin-antitoxin complex RelBE2 (Rv2865-2866) from Mycobacterium tuberculosis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes.
Structure, 21, 2013
|
|
4BBH
 
 | | Plasmodium vivax N-myristoyltransferase with a bound benzothiophene inhibitor | | Descriptor: | 2-oxopentadecyl-CoA, 3-methoxybenzyl 3-(piperidin-4-yloxy)-1-benzothiophene-2-carboxylate, CHLORIDE ION, ... | | Authors: | Rackham, M.D, Brannigan, J.A, Moss, D.K, Yu, Z, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of Novel and Ligand-Efficient Inhibitors of Plasmodium Falciparum and Plasmodium Vivax N-Myristoyltransferase.
J.Med.Chem., 56, 2013
|
|
7DXP
 
 | | Influenza H5N1 nucleoprotein in complex with nucleotides | | Descriptor: | 1,2-ETHANEDIOL, Nucleoprotein, RNA (5'-R(P*(OMU)P*(OMU)P*(OMU)P*(OMU))-3') | | Authors: | Tang, Y.S, Xu, S, Chen, Y.W, Wang, J.H, Shaw, P.C. | | Deposit date: | 2021-01-19 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of influenza nucleoprotein complexed with nucleic acid provide insights into the mechanism of RNA interaction.
Nucleic Acids Res., 49, 2021
|
|
3K98
 
 | | HSP90 N-terminal domain in complex with (1R)-2-(5-chloro-2,4-dihydroxybenzoyl)-N-ethylisoindoline-1-carboxamide | | Descriptor: | (1R)-2-[(5-chloro-2,4-dihydroxyphenyl)carbonyl]-N-ethyl-2,3-dihydro-1H-isoindole-1-carboxamide, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Gajiwala, K.S, Davies II, J.F. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dihydroxyphenylisoindoline amides as orally bioavailable inhibitors of the heat shock protein 90 (hsp90) molecular chaperone.
J.Med.Chem., 53, 2010
|
|
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
5QCZ
 
 | | Crystal structure of BACE complex with BMC015 | | Descriptor: | (4S)-4-{(S)-hydroxy[(3R,6R)-6-(methoxymethyl)morpholin-3-yl]methyl}-19-(methoxymethyl)-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDE
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000740a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N}-(4-fluorophenyl)-4-methyl-piperazine-1-carboxamide | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QDK
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000069a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, [4-(1H-benzimidazol-1-yl)phenyl]methanol | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5TY1
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-(2,4-dimethoxyphenyl)-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, SODIUM ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5D48
 
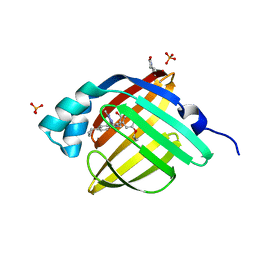 | | Crystal Structure of FABP4 in complex with 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy) phenyl]-1H-indol-1-yl}propanoic acid | | Descriptor: | 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy)phenyl]-1H-indol-1-yl}propanoic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5UD9
 
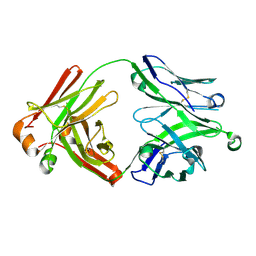 | | Crystal structure of 354BG18 Fab | | Descriptor: | Fab heavy chain, Light chain | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2016-12-23 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
5QFS
 
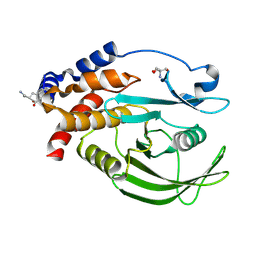 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000293a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QGB
 
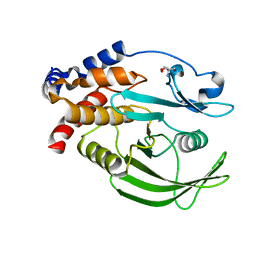 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000628a | | Descriptor: | (4R,4aR,6R,8aR)-1-benzyloctahydro-2H-6,4-(epiminomethano)-3,1-benzoxazin-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5CWC
 
 | |
4RFU
 
 | | Crystal structure of truncated P-domain from Grouper nervous necrosis virus capsid protein at 1.2A | | Descriptor: | CALCIUM ION, Coat protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
