7HED
 
 | |
7HF0
 
 | |
7HE2
 
 | |
3TZS
 
 | |
5T98
 
 | | Crystal structure of BuGH2Awt | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside Hydrolase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
7HCS
 
 | |
4HGS
 
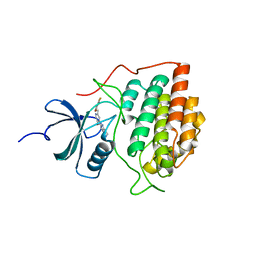 | | Crystal structure of ck1gs with compound 13 | | Descriptor: | 2-{2-[(3,4-difluorophenoxy)methyl]-5-methoxypyridin-4-yl}-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform gamma-3 | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ck1gs with compound 13
Acs Med.Chem.Lett
|
|
7HE7
 
 | |
3ZP8
 
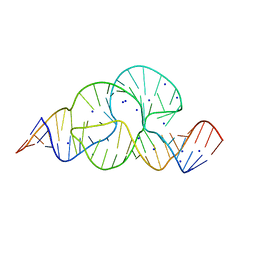 | | HIGH-RESOLUTION FULL-LENGTH HAMMERHEAD RIBOZYME | | Descriptor: | HAMMERHEAD RIBOZYME, ENZYME STRAND, SUBSTRATE STRAND, ... | | Authors: | Anderson, M, Schultz, E, Martick, M, Scott, W.G. | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Active-Site Monovalent Cations Revealed in a 1.55 A Resolution Hammerhead Ribozyme Structure
J.Mol.Biol., 425, 2013
|
|
7HFB
 
 | |
7HDS
 
 | |
7HDG
 
 | |
5TA1
 
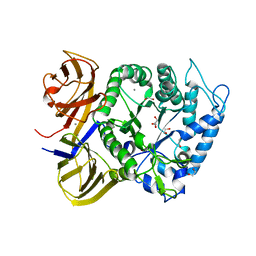 | | Crystal structure of BuGH86wt | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
7HFD
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0004127 | | Descriptor: | 2-[(1S,3aR,7aS)-2-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)octahydro-1H-isoindol-1-yl]propan-2-ol, 2-[(1S,3aS,7aR)-2-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)octahydro-1H-isoindol-1-yl]propan-2-ol, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
7HEN
 
 | |
1H1N
 
 | |
7HES
 
 | |
8DLJ
 
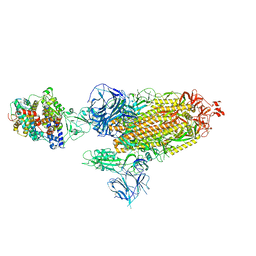 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
5D5C
 
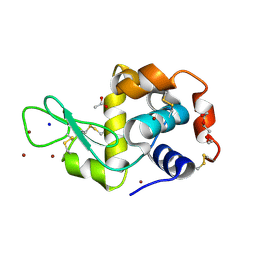 | | In meso in situ serial X-ray crystallography structure of lysozyme at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7HFR
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0005997 | | Descriptor: | (3aS,6S,6aR)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)octahydrocyclopenta[b]pyrrol-6-ol, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
5QAS
 
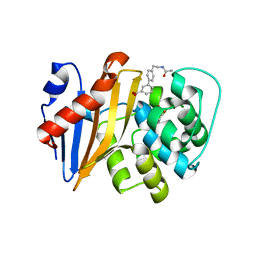 | | OXA-48 IN COMPLEX WITH COMPOUND 23b | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-(2-acetamidoethyl)phenyl]benzoic acid, Beta-lactamase | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
7HDM
 
 | |
7HE5
 
 | |
7HFC
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0004126 | | Descriptor: | 2-[(2R)-3,3-dimethyl-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-2-yl]propan-2-ol, 2-[(2S)-3,3-dimethyl-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-2-yl]propan-2-ol, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
3G23
 
 | |
