1UO1
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UO2
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UNZ
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UNX
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UNT
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
6C0F
 
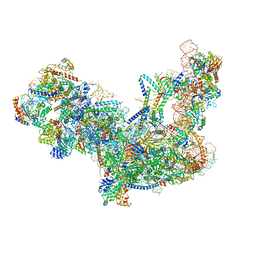 | | Yeast nucleolar pre-60S ribosomal subunit (state 2) | | Descriptor: | 5.8S rRNA, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2017-12-29 | | Release date: | 2018-03-14 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
2RB2
 
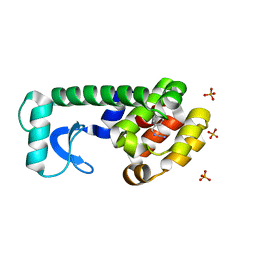 | |
2RBR
 
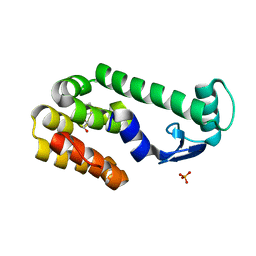 | |
2RBS
 
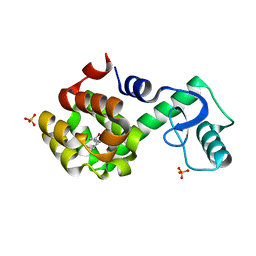 | |
2RAZ
 
 | |
2RBP
 
 | |
2RB0
 
 | | 2,6-difluorobenzylbromide complex with T4 lysozyme L99A | | Descriptor: | 2-(bromomethyl)-1,3-difluorobenzene, Lysozyme, PHOSPHATE ION | | Authors: | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | Deposit date: | 2007-09-17 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
2RBQ
 
 | |
2RBO
 
 | |
2RAY
 
 | |
2RBN
 
 | |
2WG6
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4, P61A Mutant | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2WG5
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4 | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2WQ2
 
 | |
2WQ3
 
 | |
2WQ1
 
 | |
2WPY
 
 | | GCN4 leucine zipper mutant with one VxxNxxx motif coordinating chloride | | Descriptor: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Zeth, K, Hartmann, M.D, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WPZ
 
 | | GCN4 leucine zipper mutant with two VxxNxxx motifs coordinating chloride | | Descriptor: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Zeth, K, Hartmann, M.D, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WQ0
 
 | |
5IDQ
 
 | |
