2MV8
 
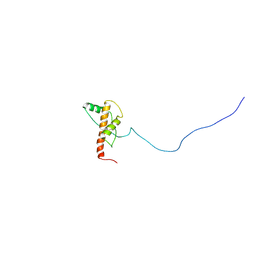 | | Solution structure of Ovis Aries PrP with mutation delta190-197 | | Descriptor: | Major prion protein | | Authors: | Munoz, C, Egalon, A, Beringue, V, Rezaei, H, Dron, M, Sizun, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Generating Bona Fide Mammalian Prions with Internal Deletions.
J.Virol., 90, 2016
|
|
1KGN
 
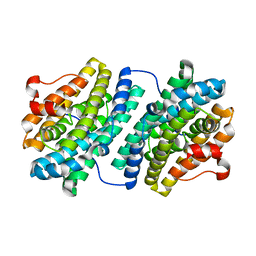 | | R2F from Corynebacterium Ammoniagenes in its oxidised, Fe containing, form | | Descriptor: | FE (III) ION, Ribonucleotide reductase protein R2F | | Authors: | Hogbom, M, Huque, Y, Sjoberg, B.M, Nordlund, P. | | Deposit date: | 2001-11-28 | | Release date: | 2001-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the di-iron/radical protein of ribonucleotide reductase from Corynebacterium ammoniagenes.
Biochemistry, 41, 2002
|
|
1KKF
 
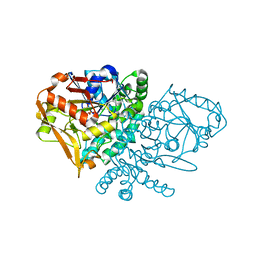 | | Complex of E. coli Adenylosuccinate Synthetase with IMP, Hadacidin, Pyrophosphate, and Mg | | Descriptor: | Adenylosuccinate Synthetase, DIPHOSPHATE, HADACIDIN, ... | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-07 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
1PS6
 
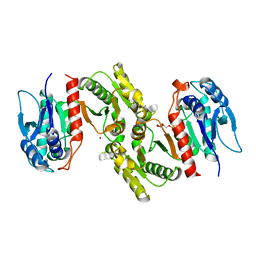 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-HYDROXY-L-THREONINE-5-MONOPHOSPHATE, 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
2N4N
 
 | |
1KJQ
 
 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-ADP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-05 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1Q2N
 
 | |
2MV9
 
 | | Solution structure of Ovis Aries PrP with mutation delta193-196 | | Descriptor: | Major prion protein | | Authors: | Munoz, C, Egalon, A, Beringue, V, Rezaei, H, Dron, M, Sizun, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Generating Bona Fide Mammalian Prions with Internal Deletions.
J.Virol., 90, 2016
|
|
7TIO
 
 | |
7TIR
 
 | |
7TIP
 
 | |
7TIS
 
 | |
7TIQ
 
 | |
4CL3
 
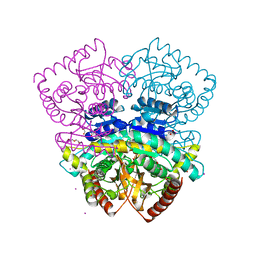 | | 1.70 A resolution structure of the malate dehydrogenase from Chloroflexus aurantiacus | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | An Experimental Point of View on Hydration/Solvation in Halophilic Proteins.
Front.Microbiol., 5, 2014
|
|
2N7O
 
 | | NMR Structure of Peptide PG-990 in DPC micelles | | Descriptor: | Peptide PG-990 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
1PMV
 
 | | The structure of JNK3 in complex with a dihydroanthrapyrazole inhibitor | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, Mitogen-activated protein kinase 10 | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | Deposit date: | 2003-06-11 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
4CQA
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase (DHODH) in complex with IDI-6273 | | Descriptor: | 2-(4-chlorobenzyl)-8-ethoxy-1,3-dimethylcyclohepta[c]pyrrol-4(2H)-one, DIHYDROOROTATE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Rowland, P. | | Deposit date: | 2014-02-12 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | In Vitro Resistance Selections for Plasmodium Falciparum Dihydroorotate Dehydrogenase Inhibitors Give Mutants with Multiple Point Mutations in the Drug-Binding Site and Altered Growth.
J.Biol.Chem., 289, 2014
|
|
1PS7
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N6H
 
 | |
2NDM
 
 | | NMR solution structure of PawS Derived Peptide 21 (PDP-21) | | Descriptor: | PawS derived peptide 21 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
2NSO
 
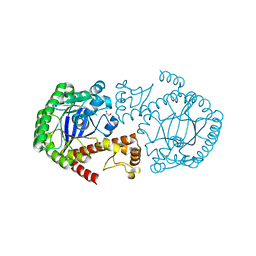 | | Trna-gunanine-transglycosylase (TGT) mutant Y106F, C158V, A232S, V233G- APO-Structure | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2006-11-06 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
4DOO
 
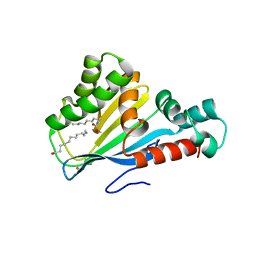 | | Crystal structure of Arabidopsis thaliana fatty-acid binding protein At3g63170 (AtFAP1) | | Descriptor: | Chalcone-flavanone isomerase family protein, LAURIC ACID, POTASSIUM ION | | Authors: | Noel, J.P, Pojer, F, Louie, G.V, Bowman, M.E. | | Deposit date: | 2012-02-09 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis.
Nature, 485, 2012
|
|
2ZGT
 
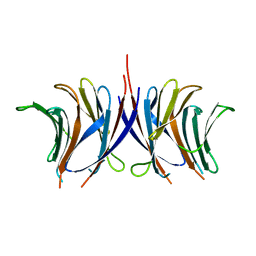 | |
1JNW
 
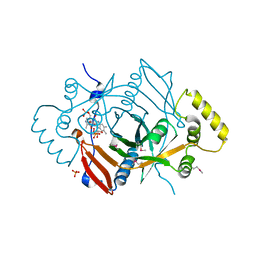 | | Active Site Structure of E. coli pyridoxine 5'-phosphate Oxidase | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | di Salvo, M.L, Ko, T.P, Musayev, F.N, Raboni, S, Schirch, V, Safo, M.K. | | Deposit date: | 2001-07-25 | | Release date: | 2001-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Active site structure and stereospecificity of Escherichia coli pyridoxine-5'-phosphate oxidase.
J.Mol.Biol., 315, 2002
|
|
