8QHH
 
 | |
8QHI
 
 | |
2STT
 
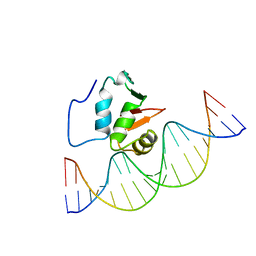 | | SOLUTION NMR STRUCTURE OF THE HUMAN ETS1/DNA COMPLEX, 25 STRUCTURES | | Descriptor: | DNA (5'-D(*TP*CP*GP*AP*AP*CP*TP*TP*CP*CP*GP*GP*CP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*CP*CP*GP*GP*AP*AP*GP*TP*TP*CP*GP*A)-3'), ETS1 | | Authors: | Clore, G.M, Werner, M.H, Gronenborn, A.M. | | Deposit date: | 1996-08-05 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Correction of the NMR structure of the ETS1/DNA complex.
J.Biomol.NMR, 10, 1997
|
|
2STW
 
 | | SOLUTION NMR STRUCTURE OF THE HUMAN ETS1/DNA COMPLEX, RESTRAINED REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*CP*GP*AP*AP*CP*TP*TP*CP*CP*GP*GP*CP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*CP*CP*GP*GP*AP*AP*GP*TP*TP*CP*GP*A)-3'), ETS1 | | Authors: | Clore, G.M, Werner, M.H, Gronenborn, A.M. | | Deposit date: | 1996-08-05 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Correction of the NMR structure of the ETS1/DNA complex.
J.Biomol.NMR, 10, 1997
|
|
5OB4
 
 | |
1BYX
 
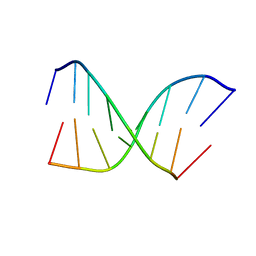 | | CHIMERIC HYBRID DUPLEX R(GCAGUGGC).R(GCCA)D(CTGC) COMPRISING THE TRNA-DNA JUNCTION FORMED DURING INITIATION OF HIV-1 REVERSE TRANSCRIPTION | | Descriptor: | DNA/RNA (5'-R(*GP*CP*CP*A)-D(P*CP*TP*GP*C)-3'), RNA (5'-R(*GP*CP*AP*GP*UP*GP*GP*C)-3') | | Authors: | Szyperski, T, Goette, M, Billeter, M, Perola, E, Cellai, L. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the chimeric hybrid duplex r(gcaguggc).r(gcca)d(CTGC) comprising the tRNA-DNA junction formed during initiation of HIV-1 reverse transcription.
J.Biomol.NMR, 13, 1999
|
|
1POG
 
 | | SOLUTION STRUCTURE OF THE OCT-1 POU-HOMEO DOMAIN DETERMINED BY NMR AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | OCT-1 POU HOMEODOMAIN DNA-BINDING PROTEIN | | Authors: | Cox, M, Van Tilborg, P.J.A, De Laat, W, Boelens, R, Van Leeuwen, H.C, Van Der Vliet, P.C, Kaptein, R. | | Deposit date: | 1994-10-12 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Oct-1 POU homeodomain determined by NMR and restrained molecular dynamics.
J.Biomol.NMR, 6, 1995
|
|
1HD6
 
 | | PHEROMONE ER-22, NMR | | Descriptor: | PHEROMONE ER-22 | | Authors: | Luginbuhl, P, Liu, A, Zerbe, O, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 2000-11-09 | | Release date: | 2000-12-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Pheromone Er-22 from Euplotes Raikovi
J.Biomol.NMR, 19, 2001
|
|
2M8W
 
 | | Restrained CS-Rosetta Solution NMR Structure of Staphylococcus aureus protein SAV1430. Northeast Structural Genomics Target ZR18. Structure determination | | Descriptor: | Uncharacterized protein | | Authors: | Mao, B, Tejero, R.T, Aramini, J.M, Snyder, D.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
2M8X
 
 | | Restrained CS-Rosetta Solution NMR structure of the CARDB domain of PF1109 from Pyrococcus furiosus. Northeast Structural Genomics Consortium target PfR193A | | Descriptor: | Uncharacterized protein | | Authors: | Mao, B, Tejero, R.T, Aramini, J.M, Snyder, D.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
2M30
 
 | | Solution NMR refinement of a metal ion bound protein using quantum mechanical/molecular mechanical and molecular dynamics methods | | Descriptor: | Repressor protein, ZINC ION | | Authors: | Chakravorty, D.K, Wang, B.I, Lee, C.I, Guerra, A.J, Giedroc, D.P, Merz Jr, K.M, Arunkumar, A.I, Pennella, M, Kong, X. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR refinement of a metal ion bound protein using metal ion inclusive restrained molecular dynamics methods.
J.Biomol.Nmr, 56, 2013
|
|
1UWD
 
 | | NMR STRUCTURE OF A PROTEIN WITH UNKNOWN FUNCTION FROM THERMOTOGA MARITIMA (TM0487), WHICH BELONGS TO THE DUF59 FAMILY. | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-02-03 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
2MUH
 
 | |
1XAX
 
 | | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae | | Descriptor: | Hypothetical UPF0054 protein HI0004 | | Authors: | Yeh, D.C, Parsons, J.F, Parsons, L.M, Liu, F, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog
Protein Sci., 14, 2005
|
|
2KBT
 
 | | Attachment of an NMR-invisible solubility enhancement tag (INSET) using a sortase-mediated protein ligation method | | Descriptor: | Proto-oncogene vav,Immunoglobulin G-binding protein G | | Authors: | Kumeta, H, Kobashigawa, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2008-12-07 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method
J.Biomol.Nmr, 43, 2009
|
|
2LEG
 
 | | Membrane protein complex DsbB-DsbA structure by joint calculations with solid-state NMR and X-ray experimental data | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein DsbA, UBIQUINONE-1, ... | | Authors: | Tang, M, Sperling, L.J, Berthold, D.A, Schwieters, C.D, Nesbitt, A.E, Nieuwkoop, A.J, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-26 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data.
J.Biomol.Nmr, 51, 2011
|
|
2KS4
 
 | |
1MAG
 
 | |
3CYS
 
 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF THE CYCLOPHILIN A-CYCLOSPORIN A COMPLEX | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Spitzfaden, C, Braun, W, Wider, G, Widmer, H, Wuthrich, K. | | Deposit date: | 1994-02-28 | | Release date: | 1994-08-31 | | Last modified: | 2017-11-01 | | Method: | SOLUTION NMR | | Cite: | Determination of the NMR Solution Structure of the Cyclophilin A-Cyclosporin a Complex.
J.Biomol.NMR, 4, 1994
|
|
2M6Q
 
 | | Refined Solution NMR Structure of Staphylococcus aureus protein SAV1430. Northeast Strucutral Genomics Consortium Target ZR18 | | Descriptor: | SAV1430 | | Authors: | Baran, M.C, Aramini, J.M, Huang, Y.J, Xiao, R, Acton, T.B, Shih, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
2ABD
 
 | |
2JQX
 
 | | Solution structure of Malate Synthase G from joint refinement against NMR and SAXS data | | Descriptor: | Malate synthase G | | Authors: | Grishaev, A, Tugarinov, V, Kay, L.E, Trewhella, J, Bax, A. | | Deposit date: | 2007-06-13 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints
J.Biomol.Nmr, 40, 2008
|
|
7ZKD
 
 | | The NMR structure of the MAX47 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, 15 N backbone and side-chain NMR assignments for three MAX effectors from Magnaporthe oryzae.
Biomol.Nmr Assign., 16, 2022
|
|
7ZJY
 
 | | The NMR structure of the MAX67 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, 15 N backbone and side-chain NMR assignments for three MAX effectors from Magnaporthe oryzae.
Biomol.Nmr Assign., 16, 2022
|
|
7ZK0
 
 | | The NMR structure of the MAX60 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, 15 N backbone and side-chain NMR assignments for three MAX effectors from Magnaporthe oryzae.
Biomol.Nmr Assign., 16, 2022
|
|
