2DCN
 
 | | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate (alpha-furanose form) | | Descriptor: | 6-O-phosphono-beta-D-psicofuranosonic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Okazaki, S, Onda, H, Suzuki, A, Kuramitsu, S, Masui, R, Yamane, T. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate
To be Published
|
|
8RCS
 
 | |
8RCM
 
 | |
6Q15
 
 | | Structure of the Salmonella SPI-1 injectisome needle complex | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
8RCL
 
 | |
6Q3G
 
 | | Structure of native bacteriophage P68 | | Descriptor: | Arstotzka protein, Head fiber protein, Inner core protein, ... | | Authors: | Dominik, H, Karel, S, Fuzik, T, Plevka, P. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
6PY9
 
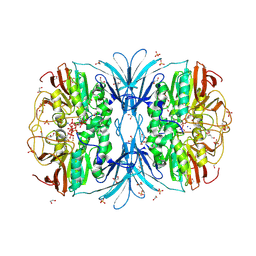 | | Crystal structure of red kidney bean purple acid phosphatase in complex with adenosine diphosphate metavanadate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADP METAVANADATE, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, McGeary, R.P, Mitic, N, Furtado, A, Schulz, B.L, Henry, R.J, Schmidt, S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elements that modulate the substrate specificity of plant purple acid phosphatases: Avenues for improved phosphorus acquisition in crops.
Plant Sci., 294, 2020
|
|
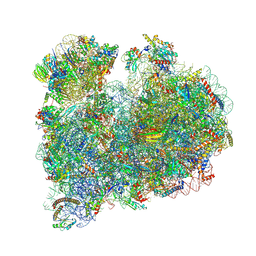
8RXX
 
 | |
8RXH
 
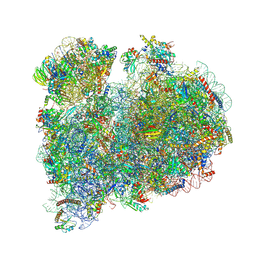 | | CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : PARENTAL STRAIN | | Descriptor: | (2S)-2-[2-[4-[[(2R,3S,4S)-3-acetyloxy-4-oxidanyl-pyrrolidin-2-yl]methyl]phenoxy]ethanoylamino]-6-azanyl-hexanoic acid, 40S ribosomal protein S12, 40S ribosomal protein S14, ... | | Authors: | Rajan, K.S, Yonath, A. | | Deposit date: | 2024-02-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification.
Cell Rep, 43, 2024
|
|
8S1P
 
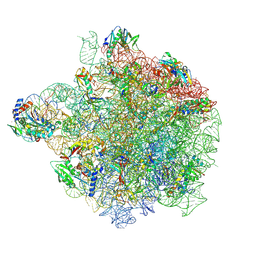 | | YlmH bound to PtRNA-50S | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2024-02-15 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | A role for the S4-domain containing protein YlmH in ribosome-associated quality control in Bacillus subtilis.
Nucleic Acids Res., 52, 2024
|
|
8R8M
 
 | |
6PWB
 
 | | Rigid body fitting of flagellin FlaB, and flagellar coiling proteins, FcpA and FcpB, into a 10 Angstrom structure of the asymmetric flagellar filament purified from Leptospira biflexa Patoc WT cells resolved via subtomogram averaging | | Descriptor: | Flagellar coiling protein A (FcpA), Flagellar coiling protein B (FcpB), Flagellin B1 (FlaB1) | | Authors: | Gibson, K.H, Sindelar, C.V, Trajtenberg, F, Buschiazzo, A, San Martin, F, Mechaly, A. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.83 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the leptospira spirochete.
Elife, 9, 2020
|
|
8R6C
 
 | |
8S6H
 
 | |
8RQE
 
 | | Composite map of bacteriophage JBD30 capsid - neck complex | | Descriptor: | Bacteriophage Mu GpT domain-containing protein, DUF1320 domain-containing protein, Mu-like prophage FluMu N-terminal domain-containing protein, ... | | Authors: | Valentova, L, Fuzik, T, Plevka, P. | | Deposit date: | 2024-01-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and replication of Pseudomonas aeruginosa phage JBD30.
Embo J., 43, 2024
|
|
6Q98
 
 | |
8S3B
 
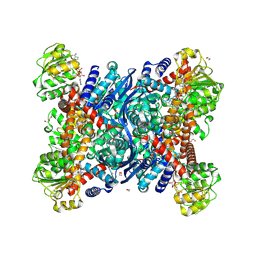 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 in complex with 3-(1H-Tetrazol-5-yl)benzoic acid and NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(1~{H}-1,2,3,4-tetrazol-5-yl)benzoic acid, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
8S8H
 
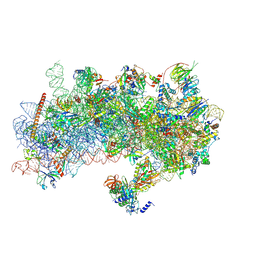 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8D
 
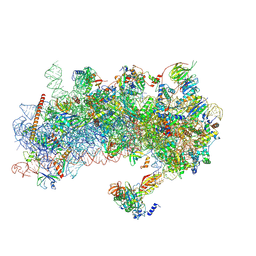 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8G
 
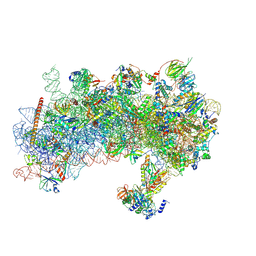 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8RW1
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8E
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8F
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8J
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF5) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8I
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
