1UYA
 
 | | THE SOLUTION STRUCTURE OF THE A-FORM OF UROGUANYLIN-16 NMR, 10 STRUCTURES | | Descriptor: | UROGUANYLIN-16, ISOMER A | | Authors: | Marx, U.C, Adermann, K, Forssmann, W.-G, Roesch, P. | | Deposit date: | 1997-09-11 | | Release date: | 1998-03-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | One peptide, two topologies: structure and interconversion dynamics of human uroguanylin isomers.
J.Pept.Res., 52, 1998
|
|
1UYB
 
 | | THE SOLUTION STRUCTURE OF THE B-FORM OF UROGUANYLIN-16 NMR, 10 STRUCTURES | | Descriptor: | UROGUANYLIN-16, ISOMER B | | Authors: | Marx, U.C, Adermann, K, Forssmann, W.-G, Roesch, P. | | Deposit date: | 1997-09-11 | | Release date: | 1998-03-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | One peptide, two topologies: structure and interconversion dynamics of human uroguanylin isomers.
J.Pept.Res., 52, 1998
|
|
1SH1
 
 | |
1S81
 
 | | PORCINE TRYPSIN WITH NO INHIBITOR BOUND | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, DeRose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1S6F
 
 | | PORCINE TRYPSIN COVALENT COMPLEX WITH BORATE AND GUANIDINE-3 INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, Derose, E.F, London, R.E. | | Deposit date: | 2004-01-23 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1TLE
 
 | | LE (LAMININ-TYPE EGF-LIKE) MODULE GIII4 IN SOLUTION AT PH 3.5 AND 290 K, NMR, 14 STRUCTURES | | Descriptor: | LAMININ | | Authors: | Baumgartner, R, Czisch, M, Mayer, U, Schl, E.P, Huber, R, Timpl, R, Holak, T.A. | | Deposit date: | 1996-01-26 | | Release date: | 1997-02-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the nidogen binding LE module of the laminin gamma1 chain in solution.
J.Mol.Biol., 257, 1996
|
|
1W4M
 
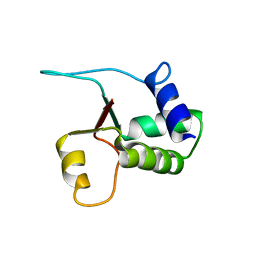 | | Structure of the human pleckstrin DEP domain by multidimensional NMR | | Descriptor: | PLECKSTRIN | | Authors: | Civera, C, Simon, B, Stier, G, Sattler, M, Macias, M.J. | | Deposit date: | 2004-07-26 | | Release date: | 2004-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Human Pleckstrin Dep Domain: Distinct Molecular Features of a Novel Dep Domain Subfamily
Proteins: Struct.,Funct., Genet., 58, 2005
|
|
1IRG
 
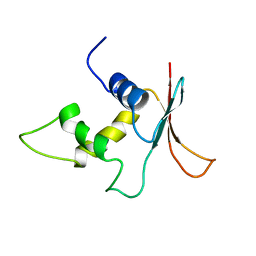 | | INTERFERON REGULATORY FACTOR-2 DNA BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | INTERFERON REGULATORY FACTOR-2 | | Authors: | Furui, J, Uegaki, K, Yamazaki, T, Shirakawa, M, Swindells, M.B, Harada, H, Taniguchi, T, Kyogoku, Y. | | Deposit date: | 1997-11-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IRF-2 DNA-binding domain: a novel subgroup of the winged helix-turn-helix family.
Structure, 6, 1998
|
|
1IRF
 
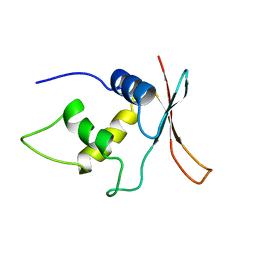 | | INTERFERON REGULATORY FACTOR-2 DNA BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERFERON REGULATORY FACTOR-2 | | Authors: | Furui, J, Uegaki, K, Yamazaki, T, Shirakawa, M, Swindells, M.B, Harada, H, Taniguchi, T, Kyogoku, Y. | | Deposit date: | 1997-11-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IRF-2 DNA-binding domain: a novel subgroup of the winged helix-turn-helix family.
Structure, 6, 1998
|
|
1IML
 
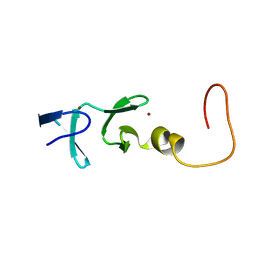 | | CYSTEINE RICH INTESTINAL PROTEIN, NMR, 48 STRUCTURES | | Descriptor: | CYSTEINE RICH INTESTINAL PROTEIN, ZINC ION | | Authors: | Perez-Alvarado, G.C, Kosa, J.L, Louis, H.A, Beckerle, M.C, Winge, D.R, Summers, M.F. | | Deposit date: | 1995-12-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the cysteine-rich intestinal protein, CRIP.
J.Mol.Biol., 257, 1996
|
|
1IMP
 
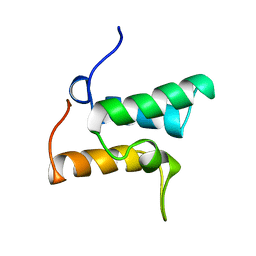 | | COLICIN E9 IMMUNITY PROTEIN IM9, NMR, 21 STRUCTURES | | Descriptor: | IM9 | | Authors: | Osborne, M.J, Breeze, A.L, Lian, L.-Y, Reilly, A, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 1996-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and 13C nuclear magnetic resonance assignments of the colicin E9 immunity protein Im9.
Biochemistry, 35, 1996
|
|
1IHV
 
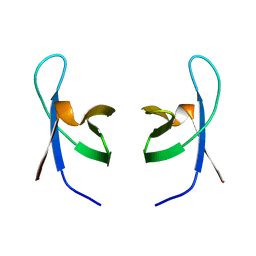 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
1IET
 
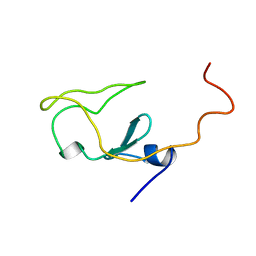 | |
1IEU
 
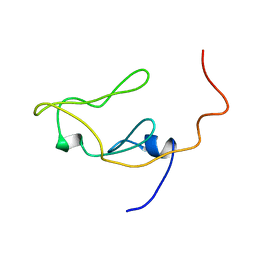 | |
1HRZ
 
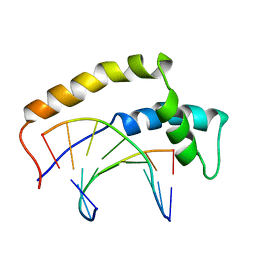 | | THE 3D STRUCTURE OF THE HUMAN SRY-DNA COMPLEX SOLVED BY MULTI-DIMENSIONAL HETERONUCLEAR-EDITED AND-FILTERED NMR | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*GP*TP*GP*C)-3'), HUMAN SRY | | Authors: | Clore, G.M, Werner, M.H, Huth, J.R, Gronenborn, A.M. | | Deposit date: | 1995-05-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex.
Cell(Cambridge,Mass.), 81, 1995
|
|
1IK4
 
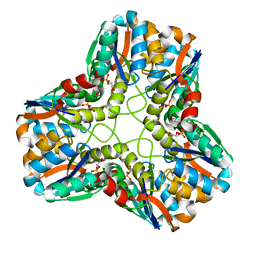 | | X-ray Structure of Methylglyoxal Synthase from E. coli Complexed with Phosphoglycolohydroxamic Acid | | Descriptor: | METHYLGLYOXAL SYNTHASE, PHOSPHOGLYCOLOHYDROXAMIC ACID | | Authors: | Marks, G.T, Harris, T.K, Massiah, M.A, Mildvan, A.S, Harrison, D.H.T. | | Deposit date: | 2001-05-02 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic implications of methylglyoxal synthase complexed with phosphoglycolohydroxamic acid as observed by X-ray crystallography and NMR spectroscopy.
Biochemistry, 40, 2001
|
|
1L5B
 
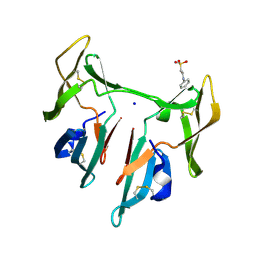 | | DOMAIN-SWAPPED CYANOVIRIN-N DIMER | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SODIUM ION, cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
1KIS
 
 | |
1IMQ
 
 | | COLICIN E9 IMMUNITY PROTEIN IM9, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | IM9 | | Authors: | Osborne, M.J, Breeze, A.L, Lian, L.Y, Reilly, A, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 1996-05-30 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and 13C nuclear magnetic resonance assignments of the colicin E9 immunity protein Im9.
Biochemistry, 35, 1996
|
|
1JLI
 
 | |
1LQI
 
 | | INSECTICIDAL ALPHA SCORPION TOXIN ISOLATED FROM THE VENOM OF SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, 29 STRUCTURES | | Descriptor: | INSECT TOXIN ALPHA | | Authors: | Tugarinov, V, Kustanovich, I, Zilberberg, N, Gurevitz, M, Anglister, J. | | Deposit date: | 1996-06-17 | | Release date: | 1997-03-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of a highly insecticidal recombinant scorpion alpha-toxin and a mutant with increased activity.
Biochemistry, 36, 1997
|
|
1LQH
 
 | | INSECTICIDAL ALPHA SCORPION TOXIN ISOLATED FROM THE VENOM OF SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INSECT TOXIN ALPHA | | Authors: | Tugarinov, V, Kustanovich, I, Zilberberg, N, Gurevitz, M, Anglister, J. | | Deposit date: | 1996-06-17 | | Release date: | 1997-03-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of a highly insecticidal recombinant scorpion alpha-toxin and a mutant with increased activity.
Biochemistry, 36, 1997
|
|
2GOV
 
 | | Solution structure of Murine p22HBP | | Descriptor: | Heme-binding protein 1 | | Authors: | Volkman, B.F, Dias, J.S, Goodfellow, B.J, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-14 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The First Structure from the SOUL/HBP Family of Heme-binding Proteins, Murine P22HBP.
J.Biol.Chem., 281, 2006
|
|
1SAA
 
 | | ATF-2 RECOGNITION SITE, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*TP*GP*AP*CP*GP*TP*CP*AP*CP*AP*TP*G)-3') | | Authors: | Conte, M.R, Lane, A.N, Bloomberg, G. | | Deposit date: | 1997-08-19 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ATF-2 recognition site and its interaction with the ATF-2 peptide.
Nucleic Acids Res., 25, 1997
|
|
1SDF
 
 | | SOLUTION STRUCTURE OF STROMAL CELL-DERIVED FACTOR-1 (SDF-1), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STROMAL CELL-DERIVED FACTOR-1 | | Authors: | Crump, M.P, Rajarathnam, K, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1997-11-15 | | Release date: | 1998-01-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1.
EMBO J., 16, 1997
|
|
