3FKH
 
 | |
3EZU
 
 | |
3FJS
 
 | |
3F9T
 
 | |
3F7X
 
 | |
3FFR
 
 | |
3GZA
 
 | |
3GWP
 
 | |
3GZB
 
 | |
3GS9
 
 | |
3HBA
 
 | |
3HBK
 
 | |
3GXG
 
 | |
3H3I
 
 | |
3H50
 
 | |
3GWR
 
 | |
3GNF
 
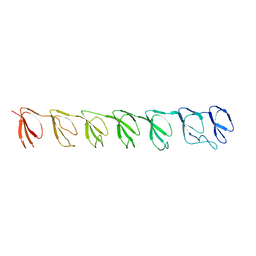 | | P1 Crystal structure of the N-terminal R1-R7 of murine MVP | | Descriptor: | Major vault protein | | Authors: | Querol-Audi, J, Casanas, A, Uson, I, Luque, D, Caston, J.R, Fita, I, Verdaguer, N. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Embo J., 28, 2009
|
|
3GXH
 
 | |
3GYA
 
 | |
3FZQ
 
 | |
3FZX
 
 | |
3G23
 
 | |
3G0K
 
 | |
3G0T
 
 | |
3GAG
 
 | |
