4WCJ
 
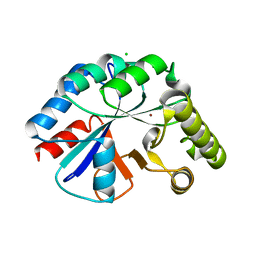 | | Structure of IcaB from Ammonifex degensii | | Descriptor: | CHLORIDE ION, Polysaccharide deacetylase, ZINC ION | | Authors: | Little, D.J, Bamford, N.C, Pokrovskaya, V, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the De-N-acetylation of Poly-beta-1,6-N-acetyl-d-glucosamine in Gram-positive Bacteria.
J.Biol.Chem., 289, 2014
|
|
4WCX
 
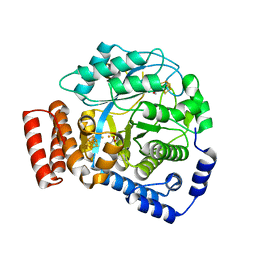 | | Crystal structure of HydG: A maturase of the [FeFe]-hydrogenase | | Descriptor: | ALANINE, Biotin and thiamin synthesis associated, FE (III) ION, ... | | Authors: | Dinis, P.C, Harmer, J.E, Driesener, R.C, Roach, P.L. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4UW2
 
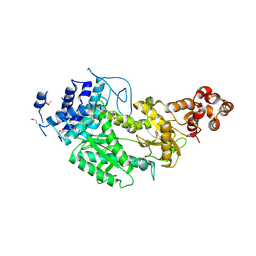 | | Crystal structure of Csm1 in T.onnurineus | | Descriptor: | CSM1 | | Authors: | Jung, T.Y, An, Y, Park, K.H, Lee, M.H, Oh, B.H, Woo, E.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Crystal Structure of the Csm1 Subunit of the Csm Complex and its Single-Stranded DNA-Specific Nuclease Activity.
Structure, 23, 2015
|
|
4URD
 
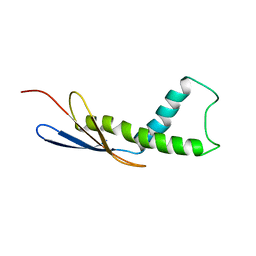 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | Descriptor: | TRIGGER FACTOR | | Authors: | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | Deposit date: | 2014-06-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
6NFK
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G bound to iodide | | Descriptor: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IODIDE ION | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
4V0S
 
 | | Crystal structure of Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3,4-DIHYDROXY-2-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]BENZOIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Santiago, C, Lamb, H, Hawkins, A.R, Maneiro, M, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-09-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
6NFP
 
 | | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | 1,2-ETHANEDIOL, Arginase, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
6NGA
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(3-(3-(dimethylamino)propyl)-2,6-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)propyl]-2,6-difluorophenyl}ethyl)-4-methylpyridin-2-amine, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
6NGK
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(3-(3-(dimethylamino)prop-1-yn-1-yl)-5-fluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)prop-1-yn-1-yl]-5-fluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
4WDX
 
 | | 17beta-HSD5 in complex with [4-(2-hydroxyethyl)piperidin-1-yl](5-methyl-1H-indol-2-yl)methanone | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [4-(2-hydroxyethyl)piperidin-1-yl](5-methyl-1H-indol-2-yl)methanone | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4USI
 
 | | Nitrogen regulatory protein PII from Chlamydomonas reinhardtii in complex with MgATP and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chellamuthu, V.R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2014-07-08 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Widespread Glutamine-Sensing Mechanism in the Plant Kingdom.
Cell(Cambridge,Mass.), 159, 2014
|
|
6NGY
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (R)-6-(2,3-difluoro-5-(2-(1-methylpyrrolidin-2-yl)ethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(2,3-difluoro-5-{2-[(2R)-1-methylpyrrolidin-2-yl]ethyl}phenyl)ethyl]-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
4USR
 
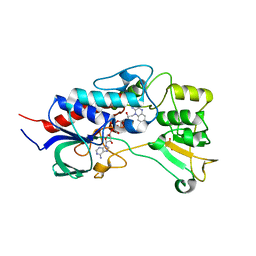 | | Structure of flavin-containing monooxygenase from Pseudomonas stutzeri NF13 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MONOOXYGENASE | | Authors: | Jensen, C.N, Ali, S.T, Allen, M.J, Grogan, G. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Exploring Nicotinamide Cofactor Promiscuity in Nad(P)H-Dependent Flavin Containing Monooxygenases (Fmos) Using Natural Variation within the Phosphate Binding Loop. Structure and Activity of Fmos from Cellvibrio Sp. Br and Pseudomonas Stutzeri NF13
J.Mol.Catal., 109, 2014
|
|
6NHE
 
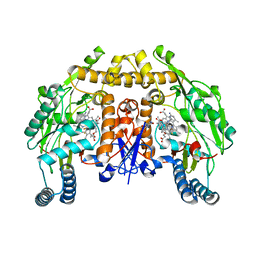 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(5-(2-((2S,4R)-4-ethoxy-1-methylpyrrolidin-2-yl)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(5-{2-[(2S,4R)-4-ethoxy-1-methylpyrrolidin-2-yl]ethyl}-2,3-difluorophenyl)ethyl]-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
4USX
 
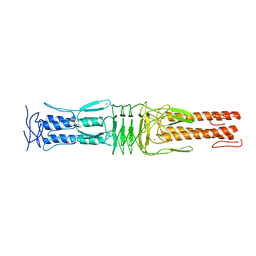 | | The Structure of the C-terminal YadA-like domain of BPSL2063 from Burkholderia pseudomallei | | Descriptor: | MAGNESIUM ION, TRIMERIC AUTOTRANSPORTER ADHESIN | | Authors: | Perletti, L, Gourlay, L.J, Peano, C, Pietrelli, A, DeBellis, G, Deantonio, C, Santoro, C, Sblattero, D, Bolognesi, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selecting Soluble/Foldable Protein Domains Through Single-Gene or Genomic Orf Filtering: Structure of the Head Domain of Burkholderia Pseudomallei Antigen Bpsl2063.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6NI4
 
 | |
6NI8
 
 | |
6NJP
 
 | | Structure of the assembled ATPase EscN in complex with its central stalk EscO from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, EscO, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6NKH
 
 | |
6ZYF
 
 | | Notum_Ghrelin complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-07-31 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Notum deacylates octanoylated ghrelin.
Mol Metab, 49, 2021
|
|
4UXL
 
 | | Structure of Human ROS1 Kinase Domain in Complex with PF-06463922 | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ROS | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2014-08-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pf-06463922 is a Potent and Selective Next-Generation Ros1/Alk Inhibitor Capable of Blocking Crizotinib-Resistant Ros1 Mutations.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6NBR
 
 | |
4USM
 
 | | WcbL complex with glycerol bound to sugar site | | Descriptor: | CHLORIDE ION, GLYCEROL, PUTATIVE SUGAR KINASE | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
6NC7
 
 | | Lipid II flippase MurJ, inward open conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, SULFATE ION | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NCZ
 
 | |
