8FP1
 
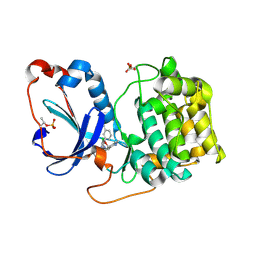 | | PKCeta kinase domain in complex with compound 2 | | Descriptor: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Protein kinase C eta type | | Authors: | Johnson, E. | | Deposit date: | 2023-01-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
3VS3
 
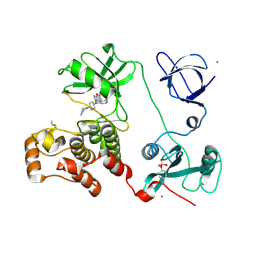 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomaebchi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS6
 
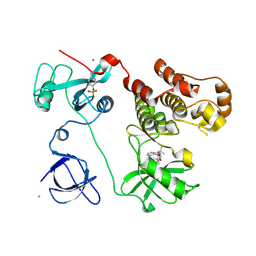 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor tert-butyl {4-[4-amino-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-2-methoxyphenyl}carbamate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein kinase HCK, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.373 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS5
 
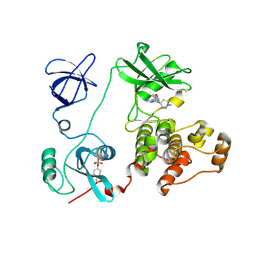 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, Tyrosine-protein kinase HCK | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3F11
 
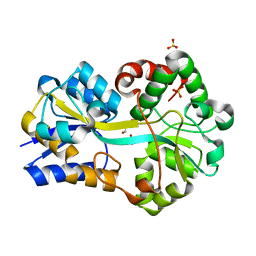 | | Structure of futa1 with iron(III) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Iron transport protein, ... | | Authors: | Koropatkin, N.M, Randich, A.M, Bhattachryya-Pakrasi, M, Pakrasi, H.B, Smith, T.J. | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the iron-binding protein, FutA1, from Synechocystis 6803
J.Biol.Chem., 282, 2007
|
|
1CR7
 
 | | PEANUT LECTIN-LACTOSE COMPLEX MONOCLINIC FORM | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-14 | | Release date: | 2001-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
2E35
 
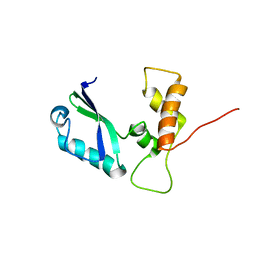 | | the minimized average structure of L11 with rg refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
7TV6
 
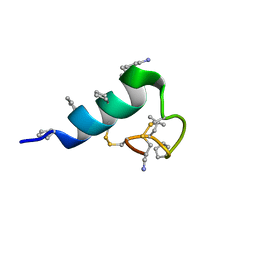 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: native loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
2E36
 
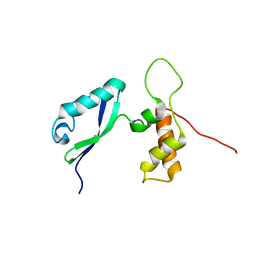 | | L11 with SANS refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
1CQ9
 
 | | PEANUT LECTIN-TRICLINIC FORM | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN) | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-06 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual
quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry
and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
6OQA
 
 | | Crystal structure of CEP250 bound to FKBP12 in the presence of FK506-like novel natural product | | Descriptor: | (3R,4E,7E,10R,11S,12R,13S,16R,17R,24aS)-11,17-dihydroxy-10,12,16-trimethyl-3-[(2R)-1-phenylbutan-2-yl]-6,9,10,11,12,13,14,15,16,17,22,23,24,24a-tetradecahydro-3H-13,17-epoxypyrido[2,1-c][1,4]oxazacyclohenicosine-1,18,19(21H)-trione, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Lee, S.-J, Shigdel, U.K, Townson, S.A, Verdine, G.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genomic discovery of an evolutionarily programmed modality for small-molecule targeting of an intractable protein surface.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2E34
 
 | | L11 structure with RDC and RG refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
2HD7
 
 | | Solution structure of C-teminal domain of twinfilin-1. | | Descriptor: | Twinfilin-1 | | Authors: | Hellman, M.H, Paavilainen, V.O, Annila, A, Lappalainen, P, Permi, P.I. | | Deposit date: | 2006-06-20 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis and evolutionary origin of actin filament capping by twinfilin
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5FLK
 
 | |
1DIV
 
 | | RIBOSOMAL PROTEIN L9 | | Descriptor: | RIBOSOMAL PROTEIN L9 | | Authors: | Hoffman, D.W, Cameron, C, Davies, C, Gerchman, S.E, Kycia, J.H, Porter, S, Ramakrishnan, V, White, S.W. | | Deposit date: | 1996-07-02 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein.
EMBO J., 13, 1994
|
|
7TV5
 
 | | Disulfide-rich venom peptide lasiocepsin: P20A mutant | | Descriptor: | Lasiocepsin | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV8
 
 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: D-Ala modified loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV7
 
 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: beta-3-Lys modified loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
5FDY
 
 | |
1DOK
 
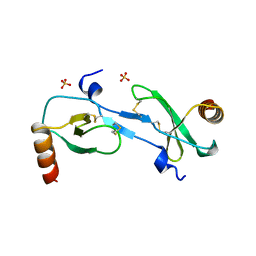 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, P-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1, SULFATE ION | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A, Domaille, P.J, Handel, T.M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1DOL
 
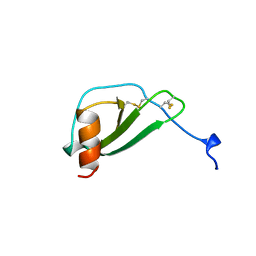 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, I-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1 | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A. | | Deposit date: | 1996-11-22 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
5UUN
 
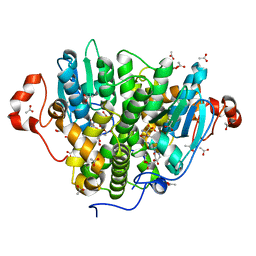 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase-like protein | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
1H7V
 
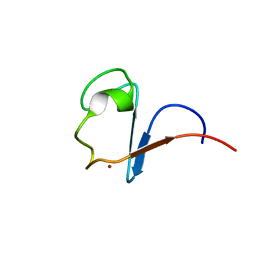 | | Rubredoxin from Guillardia Theta | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Schweimer, K, Hoffmann, S, Wastl, J, Maier, U.G, Roesch, P, Sticht, H. | | Deposit date: | 2001-01-16 | | Release date: | 2002-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a zinc substituted eukaryotic rubredoxin from the cryptomonad alga Guillardia theta.
Protein Sci., 9, 2000
|
|
6IYL
 
 | | The structure of EntE with 3-cyanobenzoyl adenylate analog | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase component of enterobactin synthase multienzyme complex, 5'-O-[(3-cyanobenzene-1-carbonyl)sulfamoyl]adenosine | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | An Engineered Aryl Acid Adenylation Domain with an Enlarged Substrate Binding Pocket.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
3ZEU
 
 | | Structure of a Salmonella typhimurium YgjD-YeaZ heterodimer bound to ATPgammaS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Nichols, C.E, Lamb, H.K, Thompson, P, El Omari, K, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-20 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Crystal Structure of the Dimer of Two Essential Salmonella Typhimurium Proteins, Ygjd & Yeaz and Calorimetric Evidence for the Formation of a Ternary Ygjd-Yeaz-Yjee Complex.
Protein Sci., 22, 2013
|
|
